Introduction
Since SARS-CoV-2 infection was first detected in Argentina in March 2020, the virus dissemination was exponential, pro-ducing a large number of cases8. The rocketing number of COVID-19 cases, which led to the occurrence of three epidemic waves (up to date), highlighted the critical role that diagnostic tests play in medical and public health decision-making to contain and mitigate the SARS-CoV-2 pandemic8. Because Argentina is a federal country, the decisions to fight the pandemic were made by each of the provinces in concor-dance with the national measures.
In Argentina, as in many other countries, the National Administration of Medicines, Food and Medical Technology granted emergency authorizations for diagnostic tests as they became available and the reference laborato-ries successively evaluated and implemented them as far as possible, according to accessibility, costs, utility and performance9. To deal with supply chain difficulties that caused shortage of reagents and equipment, clinical labo-ratories implemented multiple assay tests for SARS-CoV-2 detection in accordance with the availability of each region.
It is important to update and clearly understand the analytical parameters of the available options for SARS-CoV-2 detection, to guide the selection of tests based on certain considerations of the changing epidemiological scenario, such as expanded testing and greater access to them, introduction of vaccines, circulation of new variants, and entry of new tests, among others. With regard to the molecular methods, the licensed platforms use a variety of different primer and probe sets, diverse amplification gene targets, operational and runtime differences, equipment suggested by the manufacturer that is not always available, in addition to the variety of biological samples and extraction methods, which result in differences in analytical sensitivities that must be evaluated.
The objective of this study was to report the experi-ence in the evaluation and implementation of different tests for SARS-CoV-2 molecular detection in the central region of Argentina.
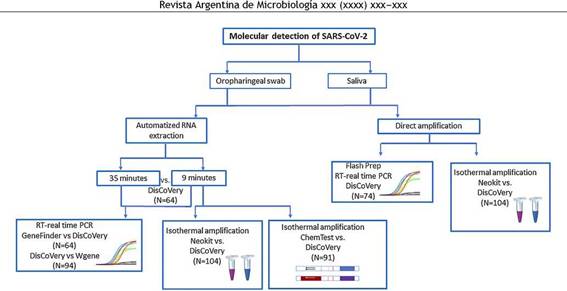
Materials and methodsDuring this study, different methods for SARS-CoV-2 molecular detection were evaluated according to the availability of kits and samples in our region at different times. The molecular assays evaluated and comparisons performed during this study are shown in Figure 1.
Figure 1: Molecular assays for SARS-CoV-2 detection tested and compared during this study.
Real time PCR
A review of the technical and operational characteristics of the following 10 RT-real time PCR kits for SARS-CoV-2 detection that were available in Argentina during the COVID-19 pandemic was carried out: ''in-house’’ coronavirus RT-real time PCR using probes and primers recommended by the CDC, LightMix Modular SARS-CoV-2 (TIB Molbiol-Syntheselabor GmbH), 2019-nCoV GENESIG (Primerdesign LTD, United Kingdom), TaqMan 2019-nCoV Assay Kit vi (Applied Biosystems, United States), GeneFinder COVID-19 Plus RealAmp Kit (Osang, Healthcare CO LTD, Korea), VIASURE SARS-CoV-2 Real Time PCR Detection Kit (Certest Biotec, Spain), Novel Coronavirus (2019-nCoV) Detection Kit v2 (Anatolia Diagnostics and Biotechnology Products Inc.), Real-Time Fluorescent RT-PCR Kit for Detecting SARS-2019-nCoV (BGI Genomics Co Ltd, China), DisCoVery SARS-CoV-2 RT-PCR Detection Kit (Safecare Biotech Hangzhou Co Ltd, China) and WGene SARS-CoV-2 RT Detection Kit (WIENER SAIC, Argentina).
We analyzed and compared the following parame-ters: target genes, internal control (CI), single reaction/ multiplex, sample volume, reagent yield, required equip-ment and reaction time.
For the comparison of GeneFinder COVID-19 Plus RealAmp Kit vs. DisCoVery SARS-CoV-2 RT-PCR Detection Kit, 64 RNA samples, previously determined for SARS-CoV-2 detection, were analyzed (positive samples n = 43; negative samples n = 21).
For the comparison of DisCoVery SARS-CoV-2 RT-PCR Detection Kit vs. WGene SARS-CoV-2 RT Detection Kit, 94 RNA samples, previously determined for SARS-CoV-2 detec-tion, were analyzed [positive samples n = 76 with Cts for the N-gene during initial diagnosis: 20 (n = 19), 20-30 (n = 29), 30 (n = 28); negative samples n = 18.
Nucleic acid extraction testFor RNA extraction, 2 automatized methods were tested: (1) MagaBio plus Virus DNA/RNA Purification Kit II (BioFlux), 35min and (2) MagaBio plus Virus RNA Purification Kit II (BioFlux), 9min, using GenePure Pro Nucleic Acid Purification System NPA-32P (Bioer). A panel of 64 positive SARS-CoV-2 oropharingeal swab samples (previously determined by RT-real time PCR DisCoVery SARS-CoV-2 RT-PCR Detection Kit) were simultaneously tested by these methods. Briefly, 300 of each sample were added to a well in a 96 well plate (one sample per well) and introduced into the equipment where the automated extraction was per-formed. For the MagaBio plus Virus DNA/RNA Purification Kit II, 10 of proteinase K were added to each well containing the sample.
FlashPrep® treatment followed by RT-real time PCRA panel of 74 oropharyngeal swab samples obtained in phys-iological saline solution, previously tested for SARS-CoV-2 detection by RT-real time PCR using DisCoVery SARS-CoV-2 RT-PCR Detection Kit [positive samples n = 58, with Cts for the N-gene during initial diagnosis: 20 (n = 4), 20-30 (n=42), 30 (n = 12); negative samples n = 16, was utilized. Briefly, 90 of each sample were incubated with 10 of the FlashPrep® RNA SARS-CoV-2 reagent (Inbio High-way, Argentina) for 15min at 55 °C and 5min at 98 °C for viral inactivation and rapid release of SARS-CoV-2 RNA, according to the manufacturer’s specifications. Then, 8 (suggested by the manufacturer) and 5 (modified according to the requirements of the real time RT-PCR kit) of the obtained RNA were used in parallel for the RT real time-PCR.
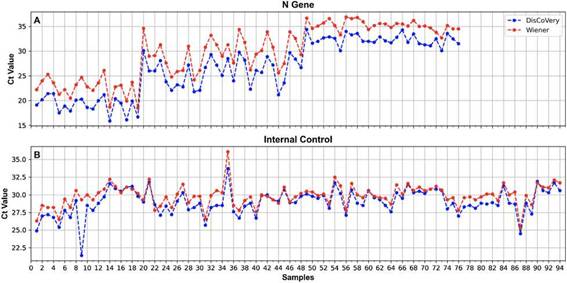
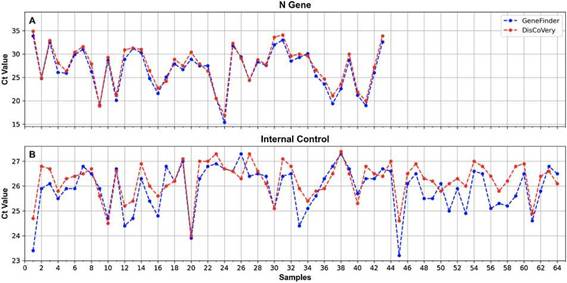 Figure 2: Ct values obtained when processing SARS-CoV-2 RNA samples using the GeneFinder (blue) and DisCoVery (red) kits for the N gene (A) and the IC (B).
Figure 2: Ct values obtained when processing SARS-CoV-2 RNA samples using the GeneFinder (blue) and DisCoVery (red) kits for the N gene (A) and the IC (B).
Isothermal amplification using Neokit Plus
A hundred and four (104) randomly selected saliva samples were processed for: 1-RNA automatized extraction with MagaBio plus Virus RNA Purification Kit II (BioFlux) 9 min, to be subjected to molecular SARS-CoV-2 detection by RT-real time PCR (using DisCoVery SARS-CoV-2 RT-PCR Detection Kit) and isothermal amplification using COVID19 Neokit Plus (Neokit S.A.S, Argenitna) (revealed with a colorimetric reaction); and 2-direct molecular detection by isothermal SARS-CoV-2 RNA amplification with COVID19 Neokit Plus (Neokit S.A.S, Argentina).
Briefly, in case No. 1 (starting from RNA), 5 of RNA of each sample were added to 35 µl of the mix provided in the kit, containing primers and enzymes, and incubated for 60 min at 64°C. In case No. 2 (starting from saliva samples, without the extraction process), 10 of each sample were added to 40 µl of a lysis buffer and incubated for 8 min at 97 °C. After that, 10 of this lysate were added to another tube with 30 of the reaction buffer and incubated for 60 min at64 °C. After that time, in both cases, No. 1 and No. 2, the colour of the samples was observed, and absorbance was measured using the provided equipment. Blue samples were considered positive, while violet samples were nega-tive. In addition, graphs of the increase in absorbance were obtained.
A reagent control, as well as positive and negative controls were included in each run.
Isothermal amplification using ELA CHEMSTRIP®A total of 91 RNA samples (extracted using MagaBio plus Virus RNA Purification Kit II 9min, as previously described), obtained from oropharyngeal swabs and previously tested for SARS-CoV-2 detection by RT-real time PCR using DisCoVery SARS-CoV-2 RT-PCR Detection Kit [positive samples n = 60, with Cts for the N-gene during initial diagnosis: 20 (n = 23), 20-30 (n = 22), 30 (n = 23); negative samples n = 23, were analyzed for isothermal amplification using an Easy Loop Amplification (ELA) and the Gen cap kit (ELA CHEMSTRIP® COVID-19, CHEMTEST ARGENTINA
S.A., Argentina), following the manufacturer’s instructions. Briefly, 5 µl of RNA were added to a mix containing Mix ELA COVID19, primers and enzyme (reagents provided by the manufacturer), and incubated for 60 min at 60 °C. Then, 300 µl of sample diluent were added, and the mixture was put onto the strip to perform immunochromatography. The result was read after 10 min. Positive and negative controls were included. An internal process control (to be added in the first step) is included in the kit.
Statistical analysesStatistical analyses were carried out using Epidat 3.1 program3. Sensitivity and specificity were calculated in most of the cases and expressed as percentages. Kappa coefficient (k) was calculated to determine how similarly the tests per-formed on a 1 -1 basis. A k of 0.8 or greater was considered a strong agreement6.
ResultsReal time PCRAfter the technical and operational revision, GeneFinder, DisCoVery and WGene were selected for further compar-isons as they exhibited the most suitable characteristics for the laboratory requirements: presence of internal control, multiplex reaction, detection of more than one gene, good performance and reduced processing time (SupplementaryTable 1).
Figure 3: Ct values obtained when processing SARS-CoV-2 RNA samples using the DisCoVery (blue) and WGene (red) kits for the N gene (A) and the IC (B).
A 100% concordance was observed when comparing GeneFinder vs. DisCoVery, as well as DisCoVery vs. WGene (k = 1 in both cases), with an average ACt value of 0.8 (range = 0-2.0) and 3.3 (range = 1.2-5.0), respectively. In the first case, Ct values were very similar (almost the same as for the N gene) (Fig. 2), and in only 2 samples the dif-ference in Cts for the N gene was as high as 2 (samples No. 4 and 12, Fig. 2A). Cts obtained for WGene were between 1.2 and 5.0 higher than those obtained by DisCoVery, mainly for the N gene (Fig. 3). Due to the aforementioned (lower Ct values), added to the fact that Discovery is less time-consuming, it was selected as the reference test for the following evaluations.
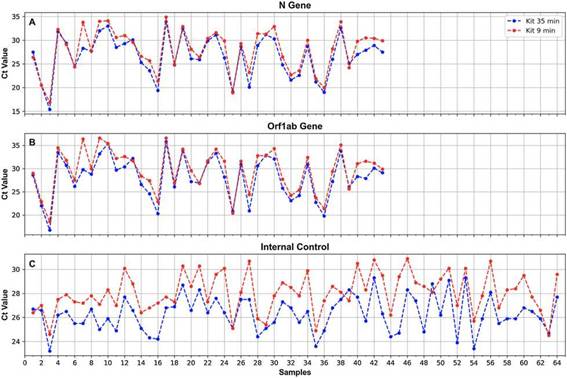
Nucleic acid extraction testsResults of the comparison of the 2 RNA extraction proto-cols showed 100% concordance for the 2 SARS-CoV-2 genes tested (N and ORF1ab) as well as the IC (all the positive samples obtained with one method were also positive with the other method) (k = 1) (Fig. 4). For both the N and the ORF1ab genes, only one sample showed a ACt as high as 5.5 (gene N) and 6.6 (ORF1ab gene) (sample No. 7, Fig. 4), with a better performance for the 35-min kit. For the rest of the samples, Cts obtained with the 35-min test for these genes were slightly lower than those obtained with the 9-min test (Fig. 4). The overall mean difference in Ct values was: for the N gene = 1.3 (range 0-5.5), for the ORF1ab gene = 1.5 (range 0-6.6).
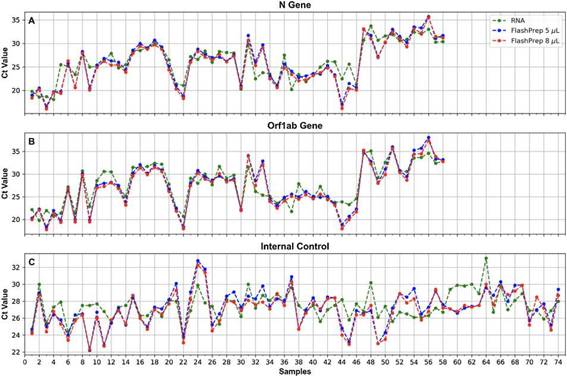
FlashPrep® treatment followed by RT-real time PCRFigure 5 shows the correlation of SARS-CoV-2 molecular detection using FlashPrep® RNA SARS-CoV-2 reagent in the 2 volumes tested (5 µl and 8 µl) vs. classical RNA extraction (using the 9-min protocol) in the 74 samples analyzed. Excel-lent correlation was observed between the results, without substantial variation of the Cts obtained using both methods, both starting from 5 µl of RNA (ACt = 0.2; range = 0.1 -5.7) as well as starting from 8 µl of RNA (ACt = 0.7, range = 0-6.1).
When comparing the results obtained using the 2 starting RNA volumes (5 vs. 8 ), a high similarity was observed in the Ct values, with a very low range between them (range = 0-1.5).
Isothermal amplification using Neokit PlusThe Neokit Plus isothermal amplification assay was evalu-ated in saliva samples, following 2 protocols: 1 - molecular detection of RNA extracted from the samples (using the 9-min protocol) and 2 - molecular detection directly from the samples (direct amplification). Both protocols were compared to RNA detection by RT real time PCR (from RNA extracted from the same samples). The sensitivity and specificity values obtained for both evaluated methods were higher than 80%, which was considered acceptable (Table 1A and Supplementary Table 2), although the Neokit Plus assay applied to SARS-CoV-2 molecular detection from RNA exhibited higher performance.
ELA CHEMSTRIP® isothermal amplificationRNA samples extracted from oropharyngeal swabs using the 9-min protocol with different Ct values, were analyzed for SARS-CoV-2 detection by ELA CHEMSTRIP® isothermal amplification and compared to results obtained by RT-real time PCR in the same samples. Sensitivity and specificity pre-sented acceptable values (higher than 80%) (Table 1B and Supplementary Table 3A).
Figure 4: Ct values obtained for the N (A) and ORF1ab (B) genes, and the IC (C), when processing oropharyngeal swab samples for SARS-CoV-2 detection using the 2 RNA extraction protocols of 35 min (blue) and 9min (red).
Table 1: Sensitivity and specificity of protocols conducted using the isothermal amplification Neokit Plus test or ELA CHEMSTRIP® compared to real time RT-PCR.
In all samples with Cts<30, 100% concordance was observed in the results obtained by ELA CHEMSTRIP® and real time PCR. However, in samples with Cts > 30, the sensitivity of the tested assay decreased considerably (Table 1B and Supplementary Table 3B).
DiscussionSome of the greatest problems of the pandemic are related to the lack of knowledge about the virus, the uncertainty and lack of resources, which pose new challenges and lead us to a constant generation of information in the field of health, including diagnosis. As the pandemic progressed, the number of COVID-19 cases increased with the identificaron of new variants, and a number of obstacles have arisen, such as the shortage of supplies (swabs, disposable material, PCR reagents, RNA isolation kits, etc.), the need for specific equipment, qualified human resources, increased testing to control and isolate cases, and the introduction of new methodologies that must be validated for all the virus variants9. This scenario raises the need to be prepared for all situations, with useful diagnostic options and strategies that contribute to the control of the out-breaks. In this context, in the reality of a developing country (such as Argentina), this work describes the experience acquired in the evaluation and implementation of different strategies and methods for the detection of SARS-CoV-2.
As in the rest of the world, SARS-CoV-2 began to be tested using in-house PCRs, with primers designed by world reference centers2. Later, commercial PCR tests became available for the amplification of different SARS-CoV-2 genomic targets, as well as isothermal tests and also new formats and reagents for RNA extraction1.
Figure 5: Ct values obtained for the N (A) and ORF1ab (B) genes, and the IC (C), when processing oropharyngeal swab samples for SARS-CoV-2 detection using FlashPrep® RNA SARS-CoV-2 reagent in the 2 volumes tested [5 pl (blue) and 8 pl (red) vs. classical RNA extraction (performed with the 9-min protocol) (green).
At the beginning of the pandemic, only 10 kits for SARS-CoV-2 detection by real time RT-PCR were available in Argentina1. Of these, GeneFinder proved to be the most reliable one, and of choice for the requirements of a lab-oratory that processed between 3000 and 5000 samples per day, such as ours. The selection was performed based on some characteristics: it had internal control, amplified more than one gene, was a multiplex reaction (all the reactions were carried out simultaneously), and achieved very good performance. After some time, the Discovery kit was intro-duced in Argentina and started to be commercialized. It exhibited similar general characteristics to GeneFinder and a high degree of concordance, but with a shorter reaction time, which is why it was selected as the gold standard for molecular diagnosis and was used for the evaluation of the rest of the methods presented here. Although the WGene kit did not show to have superior characteristics to Discovery, its performance was acceptable, therefore, it could be used in case of reagent shortage. During 2020, 2 isothermal amplification-based kits were developed and manufactured in Argentina (Neokit and ELA CHEMSTRIP®), which were tested during this study, as alternative diagnos-tic methodologies. They exhibited overall acceptable values for sensitivity and specificity (higher than 80%), proposing them as alternative diagnostic tests in places where specific infrastructure, equipment and/or trained personnel are not available. In addition, they have the advantage of low cost and short processing time (min), which allows to carry out tests on a larger scale10. However, the performance of the assays decreased when they were performed from the original sample (without RNA extraction in the case of Neokit) or for samples with low viral load (with Cts> 30), and ELA CHEMSTRIP® result was operator-dependent (since the result must be read by interpreting the presence/absence of bands in a strip), which could lead to false results. Therefore, it is always preferable to use real time RT-PCR as a first-choice technique, without ruling out the use of isothermal kits in contexts that present the aforementioned limitations.
For SARS-CoV-2 detection, nasopharyngeal (NP) spec-imens are the preferred choice samples for swab-based testing, followed by oropharyngeal (OP) specimens9; however, they must be collected by a trained healthcare worker, have an increased cross-infection risk, possibly causing discomfort, coughing and even bleeding in patients, not being so desirable for serial viral monitoring5. As an alternative option, salivary tests have demonstrated similar performance to NP and/or OP swabs for SARS-CoV-2 diagnosis4. They are a simple and non-invasive alternative, samples can be self-collected outside hospitals, without the need of a health-care professional, reducing nosocomial trans-mission risk, test waiting time, and transport and storage costs5. During this study saliva samples were tested by real time RT-PCR and isothermal amplification (direct detection and detection of RNA extracted from these samples), with acceptable performance (if the requirements for sample col-lection are met, at least 1 hour fast, no use of mouthwash or tooth brushing, among others), showing that saliva can be used for diagnosis, mainly in cases in which there is no personnel to take samples when there is a shortage of sup-plies (swabs), or in certain populations in which sampling is difficult, such as children11.
Better results were obtained using extracted RNA in the SARS-CoV-2 detection by isothermal amplification using Neokit Plus (lower performance was registered when start-ing from the original sample). On the other hand, when using the FlashPrep® RNA SARS-CoV-2 reagent, excellent results were observed for direct detection (from the original sample), even when using 5 µl as starting volume for the real time RT-PCR. The storage conditions of the samples are considered a determining factor that may have influ-enced the performance of these assays. RNA extraction has been reported to be successfully bypassed when samples are stored in a universal transport medium (UTM) or in molecular water but not when samples are stored in saline solution and in Hanks medium7. On the contrary, the FlashPrep® RNA SARS-CoV-2 reagent is prepared for the processing of sam-ples in physiological solution and does not allow the use of commercial tubes with viral transport medium. This is a factor to be considered when using this type of direct detection assays (without the extraction process), added to the need to have a thermoblock or thermal cycler for the incubation step. Despite these limitations, these techniques are useful in certain contexts, with scarce resources, lack of personnel and equipment to carry out the extraction.
Two protocols for RNA extraction with different processing times (35 and 9 min) were tested, and a high concordance in the results was observed. Although the 9-min protocol presented some differences in the ACt that could impact those cases in which the Cts are high, this difference would be negligible when evaluating other parameters that would justify its use, such as its speed, usefulness in cases of massive testing, and centralized diagnostic centres, since it allows to extract a greater number of samples in a shorter period of time, increasing the useful life of the equipment and reducing working hours Therefore, this method proved to be an excellent choice for SARS-CoV-2 RNA extraction.
ConclusiónIn conclusion, we presented an exhaustive analysis of meth-ods for molecular detection of SARS-CoV-2, showing a great variety of available assays, which can be used in different contexts and diverse epidemiological scenarios. Although the reference technique is real time RT-PCR, it requires skilled personnel and a specialized thermal cycler, tak-ing several hours to obtain results. Our results show other molecular methods or options, such as isothermal amplifica-tion or reagents to avoid RNA extraction, which have shown to be acceptable for its use in adverse contexts for rapid and accurate SARS-CoV-2 detection.
FundingThis research has not received any specific grant from agencies in the public, commercial, or non-profit sectors.
Conflict of interest
The authors declare that they have no conflicts of interest.
Acknowledgements
We thank the staff of the Laboratorio Central de la Provincia de Córdoba, for helping obtaining the samples.
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ram.2023.01.004.
Received 21 January 2022
accepted 25 January 2023












 uBio
uBio