Since the beginning of the SARS-CoV-2 pandemic, there has been a remarkable effort to find a reliable laboratory determination that could predict the severity and mortal-ity of COVID-19. Much of this attention was placed on past evidence from SARS-Coronavirus, since higher viral load was associated with worse outcomes2,8. For this purpose, a proxy for viral load called cycle threshold has been investigated in the general population.
After over 2 years of pandemic, it is widely known that the standard molecular method for COVID-19 diagnosis is via RT-PCR13. The information derived from RT-qPCR goes beyond a binary ''positive-negative’’ result. RT-qPCR mea-sures the viral RNA in terms of Ct, which is the number of cycles that the fluorescent signal requires to become detectable, and is inversely proportional to the viral load. Values >40.0 cycles are considered negative. It has been determined that respiratory samples with Ct <25.0 are considered to carry a high viral load, which was independently associated with an increased risk of death in the general population4,9.
The overall data regarding the correlation between the viral load, or the Ct, and COVID-19 severity and risk of death is controversial, whether derived from case series or sys-tematic reviews3,11,12. Several factors may affect Ct values. These have been grouped into pre-analytic (collection tech-nique, type of specimen, sampling time), analytic (internal control, type of RT-PCR used, purity of reagents, pipetting defects), and post-analytic categories (interpretation of the reports)9.
It has been demonstrated that, among oncology patients, those with hematologic malignancies have a 1.57-fold risk of experiencing severe denouements compared with patients with solid organ tumors7. Westblade et al. conducted a multicenter, observational, cohort study of patients with and without cancer. Not only did they find that those with hematologic malignancies had higher viral loads than non-cancer patients (Ct = 25.0 and 29.2, respectively), but also that the presence of a hematologic malignancy was independently associated with having a higher viral load compared with non-cancer patients (aOR = 2.52; 95% CI = 1.3-4.88; p = 0.006). Moreover, death was statistically higher in patients with a high viral load, as defined above15.
We conducted a multicenter, retrospective study with the aim of assessing whether SARS-CoV-2 Ct at diagnosis could predict mortality in patients with hematologic malignancies who contracted COVID-19. Two (2) tertiary centers in Buenos Aires participated.
Inclusion criteria: (1) Adult patients with hematologic malignancies (lymphomas, leukemias, multiple myeloma); (2) SARS-CoV-2 infection confirmed by RT-qPCR performed in either of the 2 participating tertiary centers; (3) patients admitted or treated in an out-patient basis; (4) SARS-CoV-2 infection diagnosed between March 3rd, 2020 (when the first case of COVID-19 was confirmed in Argentina) to March 31st, 2022. Exclusion criteria: incomplete data.
We extracted epidemiologic and clinical data, and the Ct value of each patient, from the medical records.
The laboratories of the participating centers used dif-ferent RT-qPCR SARS-CoV-2 kits. Thirty (30) nasal swabs from one center were analyzed using the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel (Integrated DNA Technologies - IDT, Iowa, USA), based on N1 and N2 probes for detecting SARS-CoV-2, and the human RNaseP (RP) as RNA extraction quality control. Five (5) nasal swabs from the other tertiary center were analyzed using cobas® SARS-CoV-2 Test (Roche Molecular Systems, Inc -South Branchburg, NJ, USA), which targets the ORF1ab and E genes, and uses internal control RNA (RNA IC) molecules. A mean Ct was obtained from both targeted genes in each case.
We compared the mean Ct value of the deceased patients with the survivors. We evaluated mortality due to COVID-19 rather than mortality due to the hematologic neoplasm or all-cause mortality. For those we were able to follow up
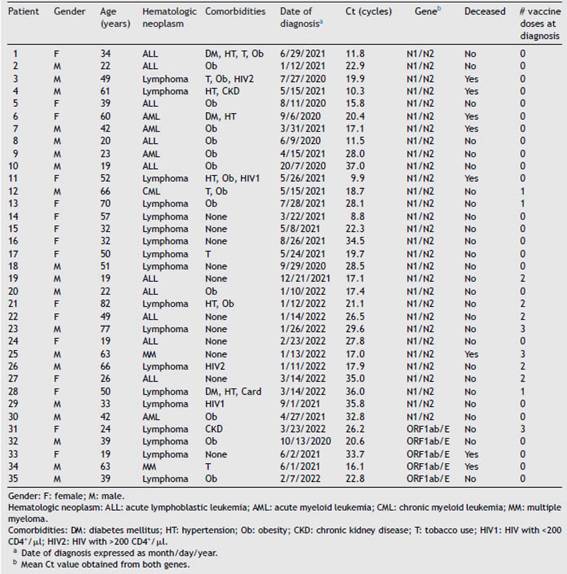
Table 1: Characteristics of SARS-CoV-2 infected patients.
Hematologic neoplasm: ALL: acute lymphoblastic leukemia; AML: acute myeloid leukemia; CML: chronic myeloid leukemia; MM: multiple myeloma.
Comorbidities: DM: diabetes mellitus; HT: hypertension; Ob: obesity; CKD: chronic kidney disease; T: tobacco use; HIV1: HIV with <200 CD4+/p,l; HIV2: HIV with >200 CD4+/µl. a Date of diagnosis expressed as month/day/year. b Mean Ct value obtained from both genes.
with sequential testing, we calculated the time to SARS-CoV-2 clearance, understood as the time elapsed between the first positive and the first negative result. The patients were not subclassified into the sequential COVID-19 waves but analyzed as a whole.
The identities of the patients included were preserved. This study was approved by the Ethics Committees of the participating institutions (Comité de Ética del Hospital de Clínicas "José de San Martín” and Comité de Ética del Sanatorio Sagrado Corazón). The procedures followed were in accordance with the Helsinki Declaration of 1975, revised in 2013 of the World Medical Association. Informed consent was obtained.
A total of 35 patients were eligible. No patient was excluded. Table 1 describes the clinical characteristics and disease denouement of the patients, their comorbidities, Ct values and vaccination status at diagnosis.
Twenty seven (27) patients (77.1%) survived, and 8 died (22.9%). Only 1 of the deceased patients had been vaccinated. Two (2) HIV+ patients died: an unvaccinated 49-year-old male with CD4+ count >200/µl and a lymphoma (patient 3), and an unvaccinated 52-year-old female - also I.M. Santarelli, D.J. Manzella, M.L. Gallo Vaulet et al.
with a lymphoma - with less than <200 CD4+/µl, among other comorbidities also shown in Table 1.
The global mean Ct was 22.8 cycles with a median of 21.7. Among the survivors, the mean Ct was 24.2, and the median was 22.9 cycles. In regard to the deceased patients, the mean Ct was 18.0 and the median Ct value was 17.0 cycles. Since we could not assume a normal distribution of Ct, we analyzed the existing difference between the survivors and deceased using the Wilcoxon Rank Sum test. This difference was significant (p = 0.035). Among the patients thatwewere able to follow-up, the mean SARS-CoV-2 time to clearance was 60 days (range = 19-158 days).
This retrospective analysis of patients with hematologic malignancies infected with SARS-CoV-2, resulted in a rel-atively small population (n = 35). One hypothesis is that uninfected cancer patients limited their hospital visits and stayed safe at home during lockdown. Some of the patients who had been diagnosed and treated for their hemato-logic malignancies could have attended other institutions for medical care. Notwithstanding this small number, we did find a statistically significant difference between the SARS-COV-2 Ct value in the nasal swabs from the patients who died, compared with those who survived.
Ct values could be useful in several clinical scenarios. First, it could help clinicians identify patients at high risk of mortality from COVID-19 at the time of diagnosis16. Sec-ond, it may serve as a guide to end isolation precautions in convalescent patients14, since Ct values above 33-34 are considered noncontagious, at least from what has been observed in vitro using Vero E6 cell cultures6. Lastly, it could help estimate the risk that could result from administering chemotherapy in patients infected with SARS-CoV-2, whose decision on whether to proceed or delay the treatment is complex due to the nature of their hematologic neoplasms. This, however, has not been universally validated and can only be used on an individual basis and with caution.
SARS-CoV-2 viral load appears to peak in the upper res-piratory tract during the first week after symptom onset, at least in immunocompetent patients1. Immunocompro-mised patients may have prolonged viral shedding and even clinical relapses5,10. In this subpopulation, which includes patients with hematologic malignancies, viral shedding has been described as long as 238 days1. In the group of patients that we were able to follow up, our results are compatible with these reports (mean 60 days, range = 19-158 days).
This study is not without limitations. First, we were not able to perform a multivariate analysis, and, therefore, our results were not adjusted for possible confounders. Of note, the COVID-19 vaccination campaign in Argentina began on December 29th, 2020. Moreover, the small sample size and the high variability of Ct could condition an artifactual result. Although Ct values are inversely proportional to the viral load, this correlation is nonlinear and should be used with caution4. It should be borne in mind that the patients’ immune status was variable and depended on numerous factors that we were not able to assess completely. The decision to compare the Ct results obtained using different RT-PCR kits was grounded on our intention to explore the clinical utility of Ct, without ignoring its limitations. In other words, when considered at the time of making a clinical decision, the gene the Ct value targets is not taken into account. We were unable to normalize the viral Ct to the internal control Ct. Finally, further studies should be conducted before adopting Ct as a universal prediction tool.
In conclusion, we found that SARS-CoV-2 Ct measured in nasal swabs obtained at diagnosis from patients with hema-tologic malignancies is associated with mortality. This may constitute an interesting line of investigation for clinical decision-making in this subpopulation.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interests
None declared.












 uBio
uBio