Introduction
Swine hemoplasmosis or swine infectious anemia, is a disease caused by the hemotropic mycoplasma My-coplasma suis and Mycoplasma parvum. These bacteria are classified within the mollicutes group and belong to the group of non-culturable hemoplasmas, adhered to the surface of porcine erythrocytes. Porcine hemoplasmosis is diagnosed on the basis of epidemiological and clinical signs, hematological and biochemical studies and by polymerase chain reaction (PCR) assay 10,23.
M. suis causes hemolytic anemia in domestic pigs producing great economic losses and is a worldwide importance disease. In Argentina, M. suis infection was first reported in the Santa Fe province in 1986, in a pig with presumptive diagnosis of pneumonía while there is no data of M. parvum infections in our country (1, 12).
M. suis infection has two forms of presentation: acute and chronic. In the former, the most important clinical signs are fever, severe anemia and death, whereas in the latter, the most important signs are mild anemia in maternity or nursery piglets, growth retardation in post-weaning and fattening pigs, reproductive failures, postpartum dysgalactia syndrome, and necrosis in the ears of sows 10,25,30.
It is pleomorphic, about 0.2 to 2.5 µm in diameter, and can occur in coccoid or rings forms. M. parvum is a non-pathogenic porcine bacterium characterized generally by the absence clinical signs. This bacterium often accumulates in large quantities in a single eryth-rocyte, however only a few red blood cells are infected. It has a size <0.5 µm 4.
Considering M. suis produce a decrease of growing rate, a higher susceptibility to secondary infections of respiratory and enteric system, production decrease and thus great economic losses 24, the aim of this study was to determine the presence of hemoplasmas in pigs from herds of Buenos Aires province, Argentina, by means of a designed nested PCR assay.
Material and methods
Design of primers to identify porcine hemoplasmas
The following hemotropic mycoplasmas 16S ribosomal gene sequences from GenBank were used for primers design: M. coccoides, M. haemocanis, M. haemofelis, M. haemomuris, M. ovis, M. wenyonii, “Candidatus M. haemominutum”, “Candidatus M. haemoparvum”, “Candidatus M. turicensis”, M. suis and M. parvum. ClustalX 2.1 software was used to build multiple alignments for each species using all the sequences so far available at GenBank 13. For each alignment, a consensus sequence with 100% inclusion threshold was created using the BioEdit program 8, so that the consensus sequences showed all the possible variants and not the most frequent one. The positions of the alignments in which the sequences presented nucleotide variability were represented in the consensus with the symbology for ambiguity of bases recommended by Cornish Bowden2. Subsequently, the consensuses of each species were aligned with ClustalX 2.1. In this last alignment, regions unique for porcine hemoplasmas were searched to design, two direct primers and two reverse primers:
MSuisEP1f 5’-AAWGGAGGCTGCCGMAAGGT-3’,
MSuisEP2f 5’-AGRTMGTTGGAGAGGTAADGGCT-3’,
MSuisEP1r 5’-CACCGCGAACACTTGTTAAGCAARTA-3’ and
MSuisEP2r 5’-AYTTTTAACAAKGRATACACAYTTCA-3’),
using Primer3 program 26,27. To include all the genotypic variants reported, the design included degenerate primers to amplify any of the possible genotypes regardless of their frequency of appearance.
Experimental design to obtain positive and negative controls o fporcine hemoplasmas
Controls were kindly provided by investigator 20 from the Institute of Porcinotecnia, Ministry of Production, Santa Fe province, Argentina. Our own controls were also obtained through a modified Groebel experimental model, based on experimental reproduction ofthe disease 7. We worked with four five-week-old females from a farm located in Buenos Aires province. Haemato-logical and PCR studies were performed to identify two of the females as positive to porcine hemoplasma and two as negative 18. Animal experimental protocols were approved by the Animal Care and Use Committee of the School of Veterinary Sciences of the National University of La Plata, Argentina (protocol number 18.1.11. res. 129/09) and the experiments were performed according to our institutional ethical rules and laws 19.
DNA extraction from porcine hemoplasmas
DNA from 100 µl of whole EDTA-blood was extracted with DNAzol (Invitrogen, USA), according to the supplier’s manual. The concentration and purity of the DNA samples were determined spectrophotometrically by measuring absorbance at 260 nm and 280 nm with a NanoVueTM (GE Healthcare, USA). DNA samples were stored at -20°C until the PCR was performed.
PCR amplification o fporcine hemoplasmas
PCR reactions contained 0.5 U/µl FIRE Pol Thermostable DNA Polymerase (Solis, Biodyne, Estonia), 1 X Buffer, 2 mM MgCl2, 0.25 mMdNTPs, 0.5 mM of each primer and nuclease-free water in 20 µl final volume. Tests were performed on a MyCycler™ thermal cycler (Bio-Rad, USA). The cycling program consisted of 2 min denaturation at 92°C, followed by 35 cycles of 30 s denaturation at 92°C, 30 s annealing at 45°C, and 40 s extension at 72°C, finishing with a 4-min extension step at 72°C. The nested PCR for swine hemoplasmas consisted of two amplification rounds, using the same conditions and cycling program described above. For the first round, 2 µl of template DNA was used, whereas for the second round, 1 µl of the first round of amplification was used as template. The second round amplification was analyzed by 1% agarose gel electro- phoresis stained with ethidium bromide (1 µg/ml). The controls mentioned above 20and our own controls were used as positive and negative controls. To detect cross-amplification with related and other species M. coccoides, M. haemocanis, M. haemofelis, M. haemomuris, M. ovis, M. wenyonii and healthy pig DNA were used. In order to detect PCR inhibitors in negative amplification samples, an internal control was used to amplify a porcine mitochondrial Cytb gene fragment using the Kocher primers describe 11.
Sequencing
PCR products were purified with polyethylene glycol according to the Travis Glennprotocol (www.mcdb. lsa.umich.edu/labs/olsen/files/PCR.pdf). The amplification products were sequenced in a MegaBACE 1000 DNA Sequencing System (GE Healthcare, USA) in both senses. Sequences were compared in GenBank through the Basic Local Alignment Search Tool (BLAST, http:// www.ncbi.nlm.nih.gov/).
Samples usedfor the standardization of the assay
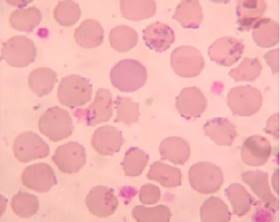
To standardize the assay, we worked with blood samples from different animal species admitted to the Central Laboratory Service of the School Hospital of the Faculty of Veterinary Sciences of the Universidad Nacional de La Plata, La Plata, Argentina, as well as from blood samples from rats and mice from the Laboratory Animal facilities of the same Faculty, 102 of which were selected because their smears stained with May Grünwald-Giemsa (Merk, USA) were positive for hemoplasma. Six of the samples were from rats, six from mice, eight from dogs, eight from cats, six from sheep, four from cattle and sixty-four from pigs. Blood samples in which coccoid forms adhered to the erythrocyte membrane were observed and which also presented alterations in the erythrocyte series (anisocytosis, poikilocytosis, polychromasia or target cell) were considered positive to hemoplasma (Figure 1).
Quality control
During the standardization stage and the samples processing, both the quality and calibration of the laboratory equipment were controlled. The analytical repeatability of the assay was estimated using different cyclers, DNA concentrations, temperatures, number of cycles, temperature gradients and commercial brands of Taq polymerase. Positive and negative reference samples were tested along with the unknown samples in each run. Throughout the study, the repeatability and precision of the assay were constantly monitored in all the samples processed.
Samples usedfor diagnosis of swine hemoplasmosis in farms of Argentina
The selection criteria for farms and pigs in each farm were based on accessibility and advisability. The study comprised 30 herds from farms of Buenos Aires province. A total 482 pig blood samples from apparently healthy animals were obtained from different categories.
Statistical analysis
An exploratory analysis of the data obtained was carried out with the EpiInfo™ 7.2.0.1 software (CDC) and InfoStat version 2018 3.
Results
For DNA extraction two alternative protocols were tested. One using silica oxide and a commercial one (DNAzol, Invitrogen®). In the comparison, with DNAzol it was necessary a lower initial blood volume and the DNA were of better quality, had smaller amount of inhibitors and result in a homogeneous yield. Because of this, DNAzol was the method of choice.
All possible combinations of primers were tested to amplify the 16S gene fragment of hemotropic mycoplasmas in a single round of PCR. As no amplification products were detected, it was necessary to perform a nested PCR by using the primers MSuisEP1f and MSuisEP1r as external primers and MSuis-EP2f and MSuis-EP2r as internal primers.
With this combination, a fragment of an approximate size of 360 bp was amplified in the second round (Figure 2). This methodology allowed to increase the sensitivity and to obtain amplification products, as well as to increase the specificity of the assay.
The amplified products, obtained from the positive controls to hemotropic mycoplasmas during the experimental reproduction of the disease, and the intensity increase of the amplified band along 45 days, are shown in Figure 3.
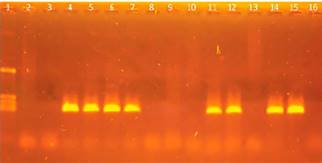
Figure 2 Nested PCR with specific primers for M.suis-M.parvum. Agarose gel electrophoresis of fragments amplified in the nested PCR with primers for Msu-is-Mparvum. The size of the amplification product was approximately 360 bp. Lane 1: marker pcDNA/HinfI (1129, 517, 453, 396 and 356 bp). Lanes 2 to 14: samples from pigs. Lane 15: positive control. Lane 16: negative control.
Lane 10 of this figure, which corresponds to day 16, shows the greatest amplification, which indi- rectly corresponds to the peak of bacteremia (Figure 3). The amplification products from the second amplification round showed a 360 pb size estimated from the molecular size marker.
The sequences of the amplification products allowed to confirm the identity of porcine hemoplasmas and to confirm that the 366-bp fragment was coincident with the expected size predicted by bioinformatics analysis.
Table 1 Sample results by MGG and PCR techniques for M.suis-Mparvum diagnosis.
| M.suis-M.parvum PCR(+) | M.suis-M.parvum PCR(-) | total | |
|---|---|---|---|
| M.suis-M.parvum MGG (+) | 72 (15%) | 82 (17%) | 154 (32%) |
| M.suis-M.parvum MGG (-) | 100 (21%) | 228 (47%) | 328 (68%) |
| total | 172 (36%) | 310 (64%) | 482 (100%) |
The amplified fragments from 10 positive samples were sequenced and the sequences compared with the GenBank database using the Blast algorithm. Products showed 100% identity to the 16S rRNA sequence described for porcine hemoplasmas.
No cross amplification was detected when M. coccoides, M. haemocanis, M. haemofelis, M. haemomuris, M. ovis, M. wenyonii and healthy pig DNA were used. The analysis of the 482 pig blood samples from the 30 farms of Argentina by using this technique showed that 36% (172/482) of the animals were positive for either M.suis or M.parvum.
The hematological studies by MGG showed a 32% (154/482) positive to porcine hemoplasmas. As have been mentioned, just a 47% of the samples analyzed by MGG were positives by PCR (Table 1).
In order to avoid false negative, an internal amplification control was used to amplify a 258 bp porcine mitochondrial DNA fragment using the Kocher et al.11 primers. All the hemoplasma negative samples amplify this control fragment confirming PCR inhibitors absence in DNA samples (Figure 4).
Discussion
In his hematological studies, Stadler y col. 23,24 from infected animals, splenectomized and no splenectomized, used a MGG stain as complementary diagnosis of anemia characterization, describing the presence of hemoplasmas as responsible of cause the disease. The hematological findings in our study agree with this author.
This technique has a low sensitivity since microorganism only are visible when there is an important peripheral blood bacterial load and subjective since are very depending of the technician experience.
Frequently leading to an under diagnosed disease. Due to the inability to cultivate hemotropic myco- plasms, reliable prevalence data for porcine hemoplasmas are rare and restricted to the post-PCR era 24.
As have been mentioned, just a 47% of the samples analyzed by MGG were positives by PCR. Although 17% were positive by MGG not by PCR, this may because this assay was designed to M.suis-M.parvum specificity it has to take into account recent studies that suggests the presence of non-classified new my- coplasma 6,22.
False positives observed in blood smears may be due these novel mycoplasmas that can be observed with MMG technique but cannot be detected by PCR due to the primer were designed to amplified M.suis- M.parvum DNA or may be due the essay subjectivity during smear reading. Though the same cannot be use as sieve test, it should be noted that is a low cost, very practical and useful test to evaluate blood cells.
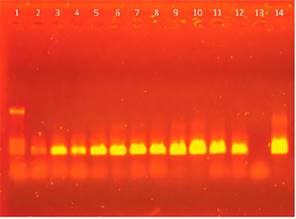
Figure 3 Nested PCR with specific primers for M.suis-M.parvum. Agarose gel elec-trophoresis of fragments amplified in the nested PCR with primers for M.suis-M parvum. The size of the amplification product was approximately 360 bp. Lane 1: marker pcDNA/HinfI (1129, 517, 453, 396 and 356 bp). Lanes 2 to 12: 0, 2, 4, 6, 8, 10, 12, 14, 16, 18 and 20 days post-surgery. Lane 13: negative control. Lane 14: positive control.
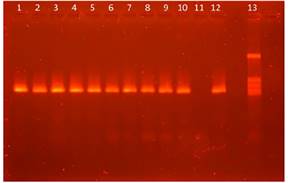
Figure 4 Agarose gel electrophoresis of the fragments amplified in the nested PCR with the primers by Kocher et al (1989). Lane 1 to 10 and 12: samples from pigs. Lane 11: negative control. Lane 12: positive control. Lane 13: marker pig mitochondrial DNA fragment.
In addition, the alterations that can be found in anemia patients are of great help for diagnosis and evolution of clinic or subclinical conditions that can be observed in animals with porcine hemoplasmosis. Although the nested PCR is a qualitative technique and can produce false negatives, these are due its detection limits.
These data cannot be compared with those from other works because is the first time that simultane- ously haematological and molecular biology studies are done in naturally infected animals by porcine hemoplasmas.
Protocols published by investigators 9,14 cannot be reproduced in our laboratory, for that reason it was necessary to design new primers to amplify porcine hemoplasma DNA. As porcine micoplasmas cannot be cultured 10,15,16,21 and since this study was carried out using a qualitative method of PCR, the sensitivity of the diagnostic test could not be determined.
The DNA sequences from the amplified fragments always showed similarity to M.suis or M.parvum. When hemotropic mycoplasma DNA obtained from a variety of species other than pig were tested, it could not be amplified confirming the specificity of the assay. The results obtained with the proper positive and negative controls in the experimental reproduction of the disease, coincided with those obtained with the controls provided 17, in the multiple amplifications carried out (Figure 3), showing high repeatability.
When the species-specific primers of M.suis were originally designed, it was thought that M.parvum and M. suis were a single species that changed shape and size as it matured 20. Other authors reported the existence of two genetic clusters among swine hemoplasmas, representing M.suis and a provisional M.parvum28.
After the genome sequencing of M. parvum in 2013, M. parvum and M. suis were classified as different species 25. Therefore, the results obtained with the primers here designed allow inferring that they amplify both M.suis and M.parvum indistinctly. The comparison of the sequences of the amplified fragment between the two species showed that there are no unique or specific nucleotide to distinguish between species.
This difference does not exist in our alignments in the rest of the 16S gene, as mentioned by other researchers 6,22. In 2012, Watanabe designed a real-time PCR by using 16 rRNA gene primers for porcine hemoplasmas. However, our alignments allowed us to observe that the specific primers for M.parvum published (29 also amplified several strains of M. suis.
This led us to conclude that their results should be revised and that those primers are not useful to differentiate strains accurately and should thus not be used. Recently was reported a novel hemoplasma species in Chinese pigs 6. Although the primers designed for M. suis or M. parvum may not amplify this species, the novel hemoplasma was not detected in pig samples from Argentina with universal primers for hemoplasmas (data not shown).
In conclusion, the results of our work showed the nested PCR here designed was a useful, repeatable and reliable technique to detect swine hemotropic mycoplasma in pigs. Sequencing confirmed that our primers always amplified M. suis or M. parvum, validating its specificity for these two species and its repeatability.
However, it would be necessary to design a new assay based on different regions of the genome to allow identifying these hemoplasmas as different species. It should also be taken into account these new hemoplasmas founded in other countries especially at the time to design new primers for the diagnosis of this bacteria in our country farms.
In addition, it is suggested to implement more exhaustive controls in cleanliness and disinfection of the establishments to reduce the presence of hematophagous vectors.
This is the first time that a nested PCR specific for porcine hemoplasmas (M. suis or M. parvum) is designed and developed in Argentina.
However, due to the advances in the knowledge of the genome of hemoplasmas worldwide, further studies on the standardization of new diagnostic assays should be conducted.