INTRODUCTION
Chromosome evolution based on ploidy, karyotype description, basic chromosome numbers and constitutive heterochromatin (C-Het) patterns is a useful tool to understand angiosperm evolutionary trajectories. Several studies have considered these data to reach phylogenetic interpretations as well as to propose character trends (Brasileiro-Vidal et al., 2007; Winterfeld & Röser, 2007; Marinho et al., 2018; Chiavegatto et al., 2019). The cytogenetic data linked to molecular data can be an effective tool to elucidate the relationships among species (Peruzzi et al.,
2009; Peruzzi & Eroğlu, 2013; Chiavegatto et al., 2020). In parallel, cytology currently proves to be useful to study species evolutionary history as well as to answer the systematic questions that arise among the different taxonomic groups of Amaryllidaceae, such as Allium L. (Khedim et al., 2016), Nothoscordum Kunth (Souza et al., 2016), Leucocoryne Lindl. (Souza et al., 2015; Sassone et al., 2018), and Ipheion Raf. (Sassone et al., 2021), all the latter being genera in which structural chromosome rearrangements and polyploidy obscure the taxonomical relationships among species.
Heterochromatin is a type of chromatin characteristic of eukaryotic chromosomes with specific functional properties that are crucial for genome stability (Janssen et al., 2018). As a morphometric chromosome marker, heterochromatin provides traceability of evolutionary trajectories of the karyotype and comprises two structurally and functionally distinct types that are termed facultative and constitutive heterochromatin (Liu et al., 2020 and references therein). Constitutive heterochromatin (C-Het), which is composed of repetitive sequences, is concentrated in the pericentromeric and telomeric domains of chromosomes (Janssen et al., 2018). The heterochromatin domain is a major, highly conserved, and structurally distinct element of eukaryotic genomes that is responsible for critical genome functions (Janssen et al., 2018). The distribution of C-Het confers a pattern of cytotaxonomic and evolutionary value to species belonging to numerous angiosperm plant groups. The staining of C-Het regions with fluorochromes, such as Chromomycin A3 [(CMA) used to distinguish chromosomal regions rich in the base pairs cytosine + guanine (CG)] and 4’6- diamidino-2-phenylindole [(DAPI) used for the localization of regions rich in the base pairs adenine + thymine (AT)], has yielded good results in comparative karyotype analyses (Schweizer, 1976). Moreover, the C-Het pattern is usually exhibited in preferential chromosomal regions, which show pattern variations ranging from interspecific differences to intraspecific polymorphisms (Lavania & Sharma, 1983; Guerra, 2000b; Almeida et al., 2022; Bacelar et al., 2023), which help to recognize karyotype evolutionary trends and taxonomic controversies. The dynamics of genome size variation is currently associated to variations in heterochromatin content, as in maize, where it has been attributed to differences in the heterochromatin located in the knobs (conspicuous heterochromatic regions) and interspersed DNA (González & Poggio, 2021). Furthermore, in angiosperms, the GC-rich repetitive DNA in the C-Het pattern frequently corresponds to C-Het that is associated to Nucleolar Organizer Regions (NORs) (Guerra, 2000b; Las Peñas et al., 2008, 2014, 2016; Acosta et al., 2016; Scaldaferro et al., 2016). Heterochromatin banding patterns can be relatively conserved at the genus level (e.g., Capsicum L.; Moscone et al., 1996) or at the family level (e.g., Cactaceae; Las Peñas et al., 2014). Further recent research has shown that environmental factors, such as latitude and altitudinal clines, are also associated to heterochromatin diversification patterns (Mata-Sucre et al., 2020; González & Poggio, 2021).
In subfamily Amaryllidoideae (Amaryllidaceae) there is no clear pattern of C-Het distribution along the karyotype, which varies both among and within genera. For example, in Galanthus L., Leucojum L., and Sternbergia Waldsein & Kitaibel, DAPI+ interstitial bands and small terminal CMA or NOR bands have been observed (D’amato & Bianchi, 1999). In the case of Crinum L., the C-Het pattern is heterogeneous. There are reports of species with DAPI+ and CMA- C-Het blocks (Ahmed et al., 2004; Alam et al., 2010) as well as species with only CMA bands at different positions (Ahmed et al., 2004). In South American species, in particular, in Placea amoena Phill, there is no AT-rich heterochromatin and the NOR regions are
located only on the short arms (Perry & Schrader, 2004). Rhodolirium laetum (Phil.) Ravenna also lacks DAPI+. heterochromatin, and 35S rDNA regions are also terminally located on the short arms (Baeza et al., 2017). Zephyranthes Herb. exhibits CMA+ bands in different patterns, either on the long arms (Z. candida (Lindl.) Herb. and Z. grandiflora Lindl.), or on the short arms (Z. citrina Baker and Z. rosea Lindl.) (Felix et al., 2011a; Daviña et al., 2022).
Zephyranthes is an American genus that includes bulb-species with ornamental and phytochemical potential and an intricated taxonomy. Currently, Zephyranthes also includes species formerly treated under Habranthus Herb., Sprekelia Heist., Haylockia Herb., Pyrolirion Herb., and Rhodophiala C. Presl (García et al., 2019). Cytologically, it presents several basic chromosome numbers, mainly x = 5, 6, 7, and 9, but most of the species studied have either the basic chromosome number x = 6 or a multiple of it (Daviña, 2001; Daviña & Honfi, 2018; Daviña et al., 2020). In genera with two or more basic chromosome numbers, the identification of the evolutionary trend to descending or ascending dysploidy is confirmed by karyomorphological information and C-Het pattern distribution (Guerra, 2000a, 2000b; 2008; Cordeiro et al., 2020), mainly because dysploidy trends can be elucidated by comparing basic karyotypes.
Diploid karyotype of Zephyranthes species based on x = 6 (2n = 2x = 12) have been found in several species, such as the endemic South American Z. andalgalensis (Ravena) S.C. Arroyo and Z. chacoensis (Ravenna) S.C. Arroyo, and also in the south American species Z. robusta (Herb.) Baker (Daviña & Honfi, 2018; Gianini Aquino et al., 2020).
On the other hand, diploids based on x = 7 have been found only three times, in Z. jamesonii (Baker) Nic. García & S.C. Arroyo (Naranjo, 1974; Di Fulvio, 1986), in Z. flavissima Ravenna (Daviña, 2001; Daviña et al., 2020) and Z. pedunculosa (Herb.) Nic. García & S.C. Arroyo (Flory, 1948; Naranjo, 1974; Daviña & Honfi, 2018; Gianini Aquino et al., 2020), all of them are South American species with restricted geographical distribution (Gianini Aquino, 2023).
In the present work, four diploid Zephyranthes species with basic chromosome numbers of x = 6 and 7 were selected in order to analyze comparatively the basic karyotype composition and distribution of C-Het in the chromosome complement. The aims of this work were i) to describe the C-Het pattern of the selected species and ii) to detect evidences that could help to understand the direction of the dysploid change among two basic chromosome numbers.
MATERIAL AND METHODS
Plant Material
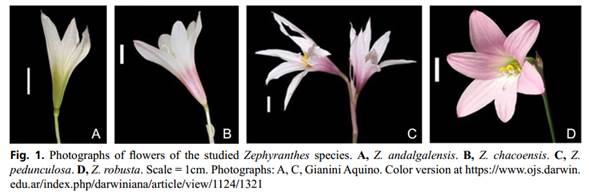
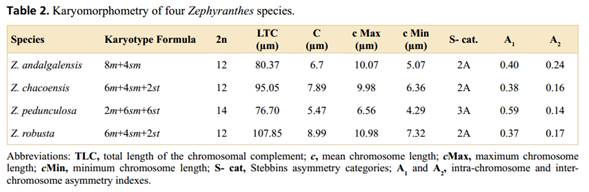
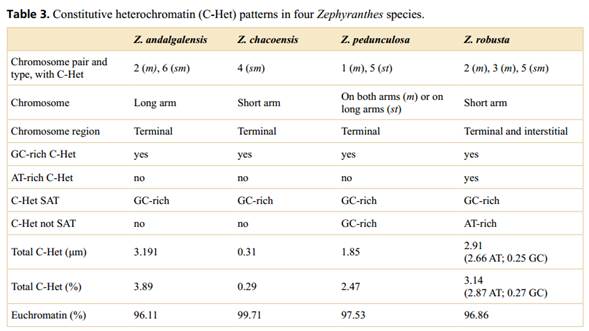
The four species studied (Figure 1, Table 1) were delimited following the descriptions proposed by Ravenna (1970), Arroyo (1990), Arroyo-Leuenberger (2009), Amaral (2011) and García et al. (2019). Plants from natural populations were collected in Argentina and cultivated in the experimental garden of the Instituto de Biología Subtropical (IBS, UNaM-CONICET), Posadas, Argentina. Voucher specimens were deposited at the Herbarium of the Universidad Nacional de Misiones (MNES). The chromosome counts of the studied materials are available in Gianini Aquino et al. (2020).

Table 1 List of the studied Zephyranthes species, locality of origin and voucher. All the specimens are stored at MNES.
Mitotic preparations
Meristem pretreatment
For cytogenetical analysis, protocols of Daviña (2001) were applied. Briefly, meristems of root tips obtained from bulbs or seeds were pretreated with 0.002M 8-hydroxyquinoline solution during 8 h at room temperature and were subsequently fixed in absolute ethanol - glacial acetic acid (3:1, v/v) and then kept in the same fixative at 4 °C for at least 72 h or until the moment of use.
Feulgen technique
Classical staining with Feulgen technique was used to observed mitotic metaphases. Root tip meristems were hydrolyzed with HCL 1N at 60 °C for 10 min and were then stained with basic fuchsin in dark chamber. Squashes were made in 2% aceto-orcein dye solution. Al least 10 cell per individual were counted.
Karyomorphometry
Karyotypes were described according to the nomenclature of Levan et al. (1964). Mean morphometric parameters were estimated based on at least 10 optimal mitotic metaphases analyzed which were stained by classical Feulgen protocol. The following parameters were used: total length of the chromosomal complement (TLC) or sum of total length of all chromosomes; mean chromosome length (c); and mean centromeric index (i). Karyotype asymmetry was analyzed using Stebbins‘ categories (1971) and Romero Zarco (1986) intra-karyotypic and inter-karyotypic asymmetry indexes (A1 and A2). Considering that the chromosome complements of a cytotype or species is diagrammatically represented as an idiogram that defines both the number and morphological features of chromosomes and that karyotypes are species specific (Honfi et al., 2017; Vimala et al., 2021), a representative species karyotype idiogram was made using Adobe Photoshop CS4 software. Satellites were classified according to Battaglia’s nomenclature (1955, 1999).
Chromosome tri-staining with fluorochromes CMA/DA/DAPI
For mitotic molecular cytogenetical analysis, protocols of Schwarzacher et al. (1980) and Daviña (2001) were applied with modifications. Each pre-treated meristem was digested in an enzyme solution (4% cellulase, 2% pectinase, in 0.01M citrate buffer, pH 4.8) for approximately 2 h and macerated in 45% acetic acid. The coverslips were removed in liquid nitrogen and the slides were dried in the air for one day at room temperature. For chromosome triple sequential staining with CMA/DA/DAPI, Schweizer (1976) protocol was applied with modifications. Slides were stained with CMA (buffer McIlvaine pH7, 10 mM MgCl2, 0.12 mg/ml chromomicin A3) in darkness for 2 h and subsequently kept in DA (distamycin) pH 7.0 MCIvaine’s buffer for 15 min. Finally, the slides were stained with DAPI (in McIlvaine buffer pH 7.0 with 1-2 μg/ml DAPI) in darkness for 30 min and aged for 3 days. Chromosomes were observed and photographed through a Leica DML epifluorescence microscope with DF C310 FX video equipment using Leica LAS V4.0 software. Chromosomal localization of C-Het in intercalary chromosome regions was determined according to index di = dx100/a of Greilhuber & Speta (1976), where d is the distance from the center of the heterochromatic band to the centromere and a corresponds to the length of the corresponding chromosome arm.
RESULTS
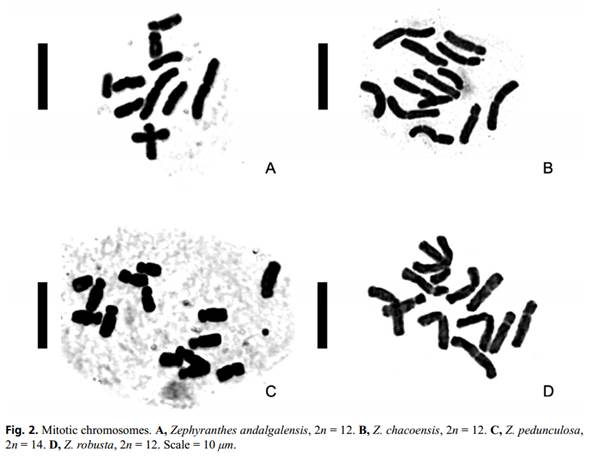
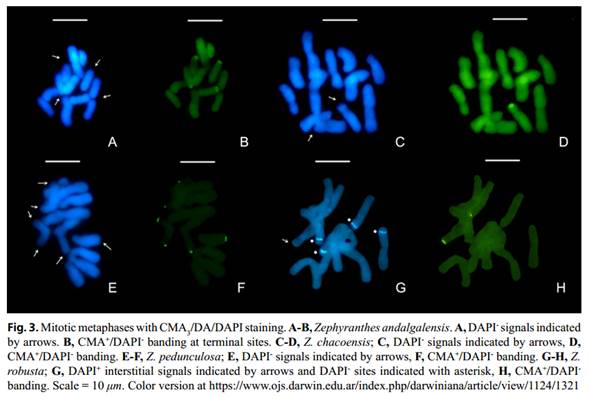
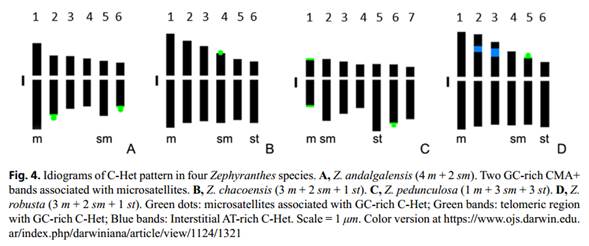
All the individuals of Z. andalgalensis were diploids with a chromosomal complement 2n = 2x = 12 and a haploid karyotype formula of 4m + 2sm (Table 2, 3, Figure 1A, 2A). Total chromosome length was found to range from 5.07 µm (±0.12 µm) to 10.07 µm (±0.23 µm). The karyotype was assigned to a 2A Stebbins’s asymmetry category (1971) and A1 = 0.40 and A2 = 0.25 following Romero Zarco (1986) indexes (Table 2). Microsatellites located at terminal position of the long arm of pairs 2 and 6 were found to be associated with CMA+/ DAPI- (Figs. 3A-B and 4A). In this species, GC-rich (CMA+/DAPI-) C-Het was found to measure 3.191 µm, thus representing 3.89% of the total chromosome length (Table3).
Zephyranthes chacoensis presented a diploid complement with 2n = 2x = 12 chromosomes and a haploid karyotype formula composed of 3m + 2sm + 1st (Fig. 2B; Table 2 and 3). Karyotype asymmetry indexes were found to belong to the 2A category of Stebbins (1971) and measured A1 = 0.38 and A2 = 0.16 (Table 3). A secondary constriction was observed on the short arm of the chromosome pair 4 (sm). The GC-rich CMA+/ DAPI- bands were found to be located on the short arms of the submetacentric pair 4 associated to the satellite (Figs. 3C-D and 4B). C-Het was GC-rich and was found to measure 0.31 µm, thus representing 0.29% of the total chromosome length of the species (Table 3).
Both accessions of Z. robusta presented a diploid complement with 2n = 2x = 12 chromosomes and a haploid karyotypic formula composed of 3m + 2sm + 1st (Table 2, 3 and Fig. 2C). The karyotype asymmetry exhibited values of A1 = 0.37 (the lowest value in all the species analyzed) and A2 = 0.17 and the assigned Stebbins (1971) category was 2A (Table 3). The highest value of TCL and chromosomal length of all the species studied was found in this species. A secondary constriction was observed on the short arm of chromosomal pair 5 (sm). The CMA+/DAPI0 bands were located on the short arm of sm pair 5 associated with the satellite, representing 0.25 µm of the total chromosome length of the species (Figs. 3H and 4D). In addition, CMA-/DAPI+ interstitial bands (AT-rich) were found on the short arms of the metacentric pairs 2 and 3 which reached 2.66 µm of the total length of the chromosome (Figs. 3G and 4D). C-Het was found to consist in AT-rich blocks (2.87% of the genome) and small GC-rich regions (0.27% of the genome) (Table 3). No polymorphism was detected among accessions.
Zephyranthes pedunculosa was observed to have a diploid complement with 2n = 2x = 14 chromosomes with a karyotype formula formed by 1 m + 3 sm + 3 st (Tables 2, 3 and Fig. 2D). The asymmetry of the karyotype resulted in A1 = 0.59 and A2 = 0.14 and the Stebbins (1971) category was 3A (Table 3). This species was observed to have the lowest value of TCL and mean chromosome length. Microsatellites located in terminal position on the long arm of subtelocentric pair 6 (st) were detected. CMA+/DAPI- bands were observed on four chromosomes (Figs. 3E-F and 4C). The metacentric pair 1 (m) was found to exhibit two terminal bands colocalized on both chromosomal arms (long and short) while in pair 6 (st), the terminal bands were associated to the microsatellite and were located on the long chromosome arm.
Abbreviations: TLC, total length of the chromosomal complement; c, mean chromosome length; cMax, maximum chromosome length; cMin, minimum chromosome length; S- cat, Stebbins asymmetry categories; A 1 and A 2 , intrachromosome and inter-chromosome asymmetry indexes.
The GC-rich (CMA+) C-Het was observed to measure 1.85 µm, thus representing 2.47% of the genome size (Table 3).
Summing up, C-Het patterns represented less than 5% of the total chromosome length in the studied diploid species (Table 3) and whereas GC-rich C-Het was found in terminal position in coincidence with microsatellites, AT-rich C-Het was rare and interstitial.
DISCUSSION
The tribe Hippeastreae (as defined in García et al., 2019) is a dysploid clade where two monophyletic subtribes were found, Traubiinae and Hippeastrinae (García et al., 2014, 2017, 2019). Hippeastrinae includes species with several basic chromosome numbers and probably a reticulate evolution caused by hybridization (García et al., 2014, 2017; Meerow et al., 2020 and references therein). Nuclear markers and plastomes suggested the existence of an Habranthus-Zephyranthes-Sprekelia polyploid complex, based on x = 6 and 7 (García et al., 2019). Some species of this complex were not studied yet, and we believe more diploids and South American endemism could contribute to a better understanding of the evolution of this group. In the latest taxonomic treatment of Hippeastreae, basic chromosome number, among other features, were used to group genera and subgenera (García et al., 2019). The basic chromosome number x = 6 chromosome is characteristic of Zephyranthes subg. Zephyranthes and Z. subg. Habranthus, and was confirmed in diploid and polyploid species, mainly tetraploids (Daviña 2001; Gianini Aquino et al. 2020; Gianini Aquino, 2023). The basic x = 7 chromosome number was found in the following diploid South American endemic species, Z. flavissima, Z. jamesonii, and Z. pedunculosa (Daviña et al., 2020).
Diverse species have uncertain basic chromosome numbers, which are probably derived from other basic numbers. Currently, few available meiotic behavior analyses, of Zephyranthes species are available (Daviña, 2001; Gianini Aquino et al., 2020, and references therein) and in consequence, the origin of polyploids remain unsolved. In line with this, meiosis has been studied only in few species to corroborate the proposed basic numbers, x = 5, 6 and 7 (Daviña & Fernández, 1989; Daviña, 2001; Daviña et al., 2020; Gianini Aquino et al., 2020).
Zephyranthes karyotypes are diverse among the species with x = 6. Diploidy seems to be the only cytological condition for Z. chacoensis (Daviña
& Honfi, 2018; Gianini Aquino et al., 2020) and all reports agree with the same karyotype formula for this species. Zephyranthes robusta is a multiploid species composed of diploid, tetraploid, hexaploid, and octoploid cytotypes (2n = 12, 24, 36, 48) and all the polyploids have an extra-American geographical distribution (Flory, 1948; Mookerjea, 1955; Nandi, 1973; Singh & Roy, 1973). Furthermore, these polyploid chromosome records belong to individuals whose origin is either unclear or indicative of materials that are under cultivation, all outside the natural geographical distribution range of the species.
We have found two basic karyotype constitutions within Zephyranthes with x = 6 chromosome number. In Z. andalgalensis, only metacentric and submetacentric chromosomes were found (4m + 2 sm) whereas in Z. chacoensis and Z. robusta, it was observed the same basic (haploid) karyotype (3m + 2sm + 1 st). Both basic haploid karyotypes, based either on x = 6 or on x= 7, share the presence of one subtelocentric pair of chromosomes. However, this conspicuous chromosome pair is not present in Z. andalgalensis. The absence of a subtelocentric pair of chromosomes in Z. andalgalensis, could be the result of structural chromosome loss or a chromosome rearrangement, e.g., an inversion. This change would have taken place before to the species dispersal in North Argentina, since currently, the geographical distribution of natural populations of northwestern are disjunct from northeastern ones. Besides, the species is endemic and in Misiones province (Argentina) lives restricted to rocky places in a small area of the Federal Reserve of Campo San Juan. The geographical isolation of the disjunct population may have facilitated the fixation of chromosome rearrangements. Future analyses of the karyotype of northwestern populations would shed light on this matter.
Although the previous reported karyotype formulas of Z. pedunculosa differ slightly from each other (Flory, 1948; sub = Habranthus juncifolius, Flory & Flagg, 1958; sub = H. juncifolius; Naranjo, 1974 sub = H. teretifolius; Daviña, 2001; Daviña & Honfi, 2018; Gianini Aquino et al., 2020), all reported 2n = 14 chromosome number for this species. The chromosomes and genome size of Z. pedunculosa are smaller than those of the other diploid species although it has an extra chromosome pair and its karyotype is more asymmetric than that of species with x = 6 chromosome numbers.
The presence of microsatellites enlarges karyotype diversification among Zephyranthes species, all of which exhibit GC -rich satellites that vary in size and location. In Z. andalgalensis, two chromosome pairs (2 and 6) have microsatellites on the long chromosome arms, this being an identity marker of the species. The same occurs in Z. pedunculosa, which was found to have a microsatellite on the long arm of the subtelocentric chromosome pair in agreement with observations reported by Naranjo (1974) and Daviña (2001). Both Z. chacoensis and Z. robusta are characterized by the presence of only one pair of chromosomes with microsatellites located on the short arm pair 4 and pair 5, respectively.
Constitutive heterochromatin content diversity can be a useful descriptor of cytotypes, ploidies and an evidence of the changes along the species evolutionary trajectories. Furthermore, C-Het is a key architectural feature of the eukaryotic chromosome. To understand karyotype evolutionary trends, the constancy and variation of chromosome constitution and patterns of heterochromatin, are very useful evolutionary clues when they are superimposed on a phylogenetic framework to elucidate the direction of chromosomal change. For example, in dysploidy (numerical modifications in basic number), heterochromatin patterns can be evidence of chromosome rearrangements and contributed to elaborate or support hypothesis about evolutionary trajectories of the karyotype.
Zephyranthes pedunculosa (x = 7) shows an additional pair of chromosomes along with a decrease in the mean chromosome length and a proper banding pattern, all of which suggests that x = 7 is a basic chromosome number derived from x = 6 species. Furthermore, diploids with 2n = 2x = 12 chromosomes of Zephyranthes have one or two GC-rich C-Het bands at terminal position per haploid complement, while diploids with x = 7 chromosomes (Z. pedunculosa) have three bands. Interestingly, Z. pedunculosa is also the only species of the genus with a conspicuous metacentric chromosome pair 1, which exhibits co-local CMA+ bands on both arms. Considering an integrative approach of cytogenetical data and a phylogenetical framework, this chromosome will help to hypothesizes about dysploid trend in this clade, for example if a similar chromosome is present in other species with x = 7 basic chromosome number, such as Z. flavissima and Z. jamesonii. Furthermore, the presence of this conspicuous metacentric chromosome in species with x = 6, becomes a potentially useful indicator to explore the ascending or descending evolutive trend, among basic chromosome numbers.
Speciation mediated via chromosome rearrangements has been documented in several angiosperms. In Clarkia Pursh (Onagraceae), the essence of speciation is chromosomal reorganization which has a significantly important role in the translocation heterozygosity that occurs in ecologically marginal sites of the geographical distribution of its prevalent related species (Lewis, 1953, 1973). In the same way, in the small South American endemic Ipheion genus (Amaryllidaceae), it has been recently proposed that the most parsimonious mechanisms that explain the current biological diversity in linages with diverse basic chromosome numbers (x = 5, 6, 7) are Robertsonian translocations (fission) and ploidy shifts (Sassone et al., 2021). Furthermore, chromosomal structural rearrangements, such as inversions and reciprocal and Robertsonian translocations, could have been involved in the evolutionary origin of the Zephyranthes basic number x = 7. In the same way, the presence of a metacentric chromosome with equilocal band pattern in both arms, also suggest that would be an isochromosome originates by mis-division, however, no meiotic behavior analyses nor x = 6-7 hybrids are available to confirm this hypothesis. Within Zephyranthes, it is easy to distinguish species using cytological characters except in the case of Z. chacoensis and Z. robusta (Barros e Silva & Guerra, 2010; Felix et al., 2011b; Gianini Aquino et al., 2020). Interestingly, in the latter two species, it is the C-Het pattern what differentiates one from the other on account of the fact that Z. chacoensis does not exhibit the DAPI+ band pattern that Z. robusta does.
In parallel, a close relationship between Z. pedunculosa and Z. robusta, was strongly suggested by a plastid signal, as well as, by ITS data (García et al., 2014, 2017). Furthermore, García et al. (2017) have considered that Z. pedunculosa present two diploid cytotypes, one with 2n = 14 chromosomes (Flory & Flagg, 1958; Naranjo, 1974) and other with 2n = 12 chromosomes (García et al., 2017). However, no evidence is currently available about the 2n = 12 cytotype of Z. pedunculosa. Numerous natural populations of Z. pedunculosa from the north of Argentina have been analyzed by chromosome counts and flow cytometry, however, no 2n = 12 cytotype, nor individuals with this number have been found (Daviña, 2001; Gianini Aquino, 2023). A plausible explanation on the origin of x = 7, considers our hypothesis that Z. pedunculosa is a derived species from a closely related x = 6 species, through a chromosomal rearrangement. In an evolutionary context, a new karyotype could be originated in a x = 6 species through a structural chromosomal change, such as Robertsonian rearrangements (complete chromosome arm fusion or fission) and then, through a subsequent fixation of the new chromosomal rearrangement in a homozygous state, the x = 7 of Zephyranthes originate. Structural chromosome changes can explain the loss of one metacentric pair and the addition of the two subtelocentric pairs in Z. pedunculosa observed in our study. Further evidence based on the meiotic behavior of synthetic interspecific hybrids and fluorescent in situ hibridization (FISH) is necessary to confirm our hypothesis.
The C-Het patterns of polyploid Zephyranthes species are also species-specific. In the polyploid complex Z. sylvatica (Mart. ex Schult. & Schult.f.) Baker, Felix et al. (2011a, 2001b) analyzed diploid and triploid individuals.
In all cases, they observed a large terminal CMA+ block per haploid complement as well as a smaller one in another chromosome pair, both located on the long arms. Felix et al. (2011b) suggest that this may be due to transposition events in the heterochromatin region, and consequently, in Z. sylvatica switches from having one signal to two signals per chromosomal complement. Furthermore, in the tetraploid cytotype of Z. brachyandra (Baker) Backer, a complex C -Het pattern with CMA+ and DAPI+ C-Het bands was reported (Nascimento et al., 2022) . The AT-rich DAPI+ C-Het blocks have been unfrequently reported in Zephyranthes species and until now they have been documented only in Z. robusta (2x) and Z. brachyandra (4x) (Fig.4; Barros e Silva & Guerra, 2010, Felix et al., 2011a, 2001b; Nascimento et al., 2022). Currently, Z. robusta, Z. martinezii (Ravenna) Nic. García, and Z. brachyandra, are phylogenetically related in a well-supported clade (García et al., 2014, 2017). In agreement with these results, our cytogenetical evidence strongly supports the close relationship proposed for Z. robusta and Z. brachyandrus. According to Daviña & Honfi (2018), meiotic behavior indicates that Z. brachyandra is an allopolyploid. Morphology, karyotypes, C-Het patterns, meiotic behavior analyses and the geographical distribution of the natural population of both species, all give support to the hypothesis that Z. robusta is one of the diploid ancestors of the allopolyploid Z. brachyandra.
Until now, it is not available the heterochromatin pattern of Z. martinezii. The synthetic interspecific allotriploid hybrids obtained by Traub (1952) is a further evidence of this relationship. Within the Hippeastreae clade, numerous interspecific crosses have been successfully performed, demonstrating, on the one hand, the permeability of the reproductive barriers among taxa (Traub, 1952; Flory, 1968; Cage, 1969; Knobloch, 1972; Flory & Smith, 1980a, 1980b; Howard, 1990; Chowdhury & Hubstenberger, 2006; David, 2011), and, on the other hand, the possibility of allopolyploid speciation within Zephyranthes species.
Summing up, in Zephyranthes species, AT-rich C-Het is rare whereas GC-rich heterochromatin is generally associated with terminal satellites. The heterochromatin patterns and the karyomorphometric features of karyotypes are useful tools to distinguish species as well as to identify differences and affinities among them. Further research within the framework of ongoing evolutionary studies will provide new insights into a larger number of Hippeastrinae species with data on C-Het patterns. To date, it is known that the diploids of x = 6 and 7, two Zephyranthes basic chromosome numbers, co-habit geographically in the subtropics of South America. Nonetheless, in order to make a robust ancestral karyotype reconstruction of the genus, which is critical for our understanding of genome evolution, is necessary to contrast a biogeographic analysis of the species.












 uBio
uBio