Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO
Links relacionados
-
 Similares en
SciELO
Similares en
SciELO
Compartir
Revista agronómica del noroeste argentino
versión impresa ISSN 0080-2069versión On-line ISSN 2314-369X
Rev. agron. noroeste arg. vol.37 no.1 San Miguel de Tucumán jun. 2017
SCIENTIFIC ARTICLE
Antagonism between Azospirillum brasilense Az39 and Pseudomonas oryzihabitans, a seed-borne endophyte, in growing rice plants
Antagonismo entre Azospirillum brasilense Az39 y Pseudomonas oryzihabitans, una endófita de semilla, en plantas de arroz
G. Rariz; L. Ferrando; N. Echegoyen; A. Fernández Scavino*
Área Microbiología, Departamento de Biociencias, Facultad de Química, Universidad de la República. General Flores 2124, Casilla de Correo 1157, (11800), Montevideo, Uruguay. *E-mail: afernand@fq.edu.uy, ana.fernandez.scavino@gmail.com
Abstract
The interactions between native endophytic bacteria and inoculated beneficial bacteria in plant tissues are relevant to guarantee the success of commercial inoculants assuming that benefitial bacteria must persist associated to the plant for a certain period of time. In this study we examine whether Pseudomonas oryzihabitans is able to antagonize Azospirillum brasilense Az39, a Plant Growth Promoting Bacteria widely used in South America. Surface-sterilized riceseeds inoculated with high amounts of P. oryzihabitans G16 or A. brasilense Az39 or with both strains were grown under hydroponic conditions. Shoot and root biomass of 15 days-old inoculated-seedlings were compared to those of uninoculated seedlings. In addition, enumeration and identification of endophytic bacteria were performed on these seedlings. Heterotrophic and siderophore-producing bacteria isolated from seedlings were identified by 16S rRNA gene partial sequencing. A. brasilense and P. oryzihabitans were able to colonize rice plants, being predominant in the roots and shoots of the respective inoculated seedlings. In co-inoculated plants, only P. oryzihabitans was recovered. The siderophore-producing Sphingomonas sp. was predominant in uninoculated plants and was detected in all inoculated plants. The vegetal biomass was the lowest in P. oryzihabitans-inoculated plants and the highest in A. brasilense inoculated plants.This work shows that P. oryzihabitans antagonizes A. brasilense in plant tissues and decreases rice plant yield. Other seed-borne endophytes, particularly siderophore-producer bacteria of the genus Sphingomonas, are not outcompeted by P. oryzihabitans.
Keywords: Plant growth promotion; Co-inoculation; Plant colonization; Siderophore production.
Resumen
Las interacciones entre las bacterias nativas endófitas y bacterias benéficas inoculadas en los tejidos vegetales son relevantes para garantizar el éxito de los inoculantes comerciales asumiendo que las bacterias benéficas deben persistir asociadas a la planta durante un cierto período de tiempo. En este estudio se analiza si Pseudomonas oryzihabitans es capaz de antagonizar Azospirillum brasilense Az39, una bacteria promotora del crecimiento vegetal ampliamente utilizada en América del Sur. Las semillas de arroz esterilizadas superficialmente, inoculadas con altas cantidades de P. oryzihabitans G16 o A. brasilense Az39 o de ambas cepas, se cultivaron en condiciones hidropónicas. Se comparó la biomasa de brotes y raíces de plántulas inoculadas con la de las plántulas no inoculadas al cabo de 15 días de crecimiento. Se cuantificaron e identificaron las bacterias endófitas presentes en esas plántulas. Las bacterias heterótrofas y productoras de sideróforos aisladas se identificaron mediante secuenciación parcial del gen 16S rRNA. A. brasilense y P. oryzihabitans fueron capaces de colonizar las plántulas de arroz, siendo predominantes en las raíces y brotes de las plántulas inoculadas con cada una de estas cepas. En plantas co-inoculadas sólo se recuperó P. rysihabitans.Las bacterias productoras de sideróforos pertenecientes al género Sphingomonas predominaron en plántulas no inoculadas y se detectaron en todas las plántulas inoculadas. Los valores más bajos de biomasa vegetal se encontraron en plántulas inoculadas con P. oryzihabitans y los más altos en plántulas inoculadas con A. brasilense. Este trabajo muestra que P. oryzihabitans antagoniza A. brasilense en tejidos vegetales y disminuye el rendimiento de la planta de arroz. Otras endófitas asociadas a semilla, particularmente las bacterias productoras de sideróforos del género Sphingomonas, no fueron antagonizadas por P. oryzihabitans.
Palabras clave: Promoción del crecimiento vegetal; Co-inoculación; Colonización de plantas; Producción de sideróforos.
Received 04/26/17; Accepted 06/23/17.
The authors declare to have no conflict of interests.
Introduction
Cultivated rice (Oryza sativa) is the most important staple crop in the world. Irrigating rice is a highly water-consuming agronomic process that affects water quality due to the use of agrochemicals. To satisfy the increasing world demand for sustainable rice production, biotechnological alternatives have been proposed. Among of them, inoculation with Plant Growth Promoting Bacteria (PGPB) is a valuable tool to improve crop yield, minimizing the use of fertilizers and pesticides. This technology has been expanded worldwide, particularly in Latin American countries (Valverde et al., 2015).
Azospirillum spp. is one of the PGPB most widely used by farmers to increase the yield of several crops. The ability of the species Azospirillum to stimulate plant growth has been attributed to the production of the auxin indole-3-acetic acid (IAA), gibberellins, polyamines and amino acids (Bashan and de-Bashan, 2010). Azospirillum brasilense and A. lipoferum have been commonly used in commercially available inoculants for a variety of crops, with strain A. brasilense Az39 being the most frequently employed in biofertilizers. Several reports indicate that inoculation with Az39 significantly improved the yields of many cereals (Fulcheri and Frioni, 1994; Cassán et al., 2009; Zawoznik et al., 2011; García de Salamone et al., 2012), although other authors did not find such auspicious results (Piccinin et al., 2011; Araújo et al., 2013; Hungria et al., 2013).
The observed variability in the crop responses to the use of Azospirillum spp. as biofertilizer can be attributed to the differences in the conditions that the strain has to overcome to be installed successfully in the plant. The plant-PGPB interaction is essential for an effective improvement in vegetal yield, and it could be influenced by biotic and abiotic parameters. Among the biotic parameters that affect the inoculation efficiency, the interaction between PGPB and the naturally occurring endophytic bacteria has rarely been studied. Endophytes are defined as microorganisms that can be isolated from surface-sterilized plant materials, inhabiting plant tissues without causing harm to them (Hallmann et al., 1997). The competition between native endophytic bacteria and inoculated bacteria may reduce the activity of PGPB and consequently diminish crop yield. Thus, the success of the endophytic PGPB and the persistence of the plant-bacteria association depends on the ability of the inoculated bacteria to grow inside the plant tissues, where native endophytes can also grow.
The main sources of endophytes are the seeds and the rhizospheric soil from where certain bacteria can be attracted towards the roots by plant exudates. Seed-borne endophytes have been examined in different rice varieties and regions (Okunishi et al., 2005; Mano et al., 2006; Loaces et al., 2011; Ruiza et al., 2011). It was shown that endophytes are able to inhibit beneficial bacteria in a dual plate antagonistic assay. Such is the case of Pseudomonas oryzihabitans, a common species detected in rice (Cottyn et al., 2009; Ruiza et al., 2011; Hardoim et al., 2012), for which at least one strain (G16) has been able to inhibit several species of Azospirillum spp. and Herbaspirillum spp. employed in commercial biofertilizers (Loaces et al., 2011). It has been suggested that inside the plants there may exist antagonistic interactions among endophytic bacteria like to those described among the rhizospheric community. (Muñoz Rojas et al., 2005). Moreover, A. brasilense was excluded from the rhizoplane of rice plantlets by endophytes of the genera Bacillus and Corynebacterium (Bacilio-Jiménez et al., 2001). However, antagonism between PGPB and endophytes into vegetal tissues,including shoots, has not been explored.
The aim of this work was to determine if the seed-borne strain Pseudomonas oryzihabitans G16 is able to antagonize A. brasilense Az39 into plant tissues of rice. The seeds were inoculated or co-inoculated with similar amounts of these strains, and 15 days-old seedlings were analysed for endophytic bacterial abundance, and the predominant isolated bacteria were further characterized. Vegetal yield parameters were compared for plants with different inoculation treatments. In addition, seeds from other rice varieties were examined to determine whether P. oryzihabitans is a ubiquitous endophyte.
Material and methods
Seeds sterilization
Three different rice (Oryza sativa) varieties were used in this study: INIA-Olimar, El Paso 144 and INIA-Tacuarí. Seeds (5 g) of each variety were peeled and hydrated in 80 mL of sterilized distilled water for one hour. The seeds were surface-sterilized by shaking in a solution of NaClO 1.7% for 5 min, followed by four, 3 min washes with 200 mL of sterile water. Sterility was tested by rolling the seeds on R2A plates, where no growth was observed after incubation at 30ºC for 48 h.
Experimental design of the plant growth assay under hydroponic conditions
The variety Olimar was used for this assay. Plants were grown in a climate-controlled chamber with 16 h of daylight at 25ºC and 80% relative humidity. A randomized complete block design with two blocks (or replicates) per treatment was used. Each replicate was composed of 14 tubes containing 2 plants per tube. The treatments consisted of seeds inoculated with: Azospirillum brasilense Az39 (Az39), Pseudomonas oryzihabitans G16 (G16), or co-inoculated with A. brasilense Az39 and P. oryzihabitans G16 (Az39+G16). Uninoculated seeds were used as control treatment (C). The blocks were distributed completely at random in the chamber.
Inoculation of seeds
Strains of A. brasilense Az39 and P. oryzihabitans G16 were grow by 18 h in Tripticase Soy Broth (TSB) at 30ºC. The inocula were diluted with fresh medium to 0.1 OD600. The bacterial density was verified by plate counting in R2A as described below. The bacterial suspensions contained per mL: 2.4 x 107 CFU (Az39), 1.3 x 108 CFU (G16) and, for the co-inoculation treatment, 2.0 x 108 CFU of Az39 and 1.0 x 108 CFU of G16. These values are in the range of the bacterial density recommended for the application of commercial products with A. brasilense AZ39 (http://www.calister.com.uy/producto/bioprom/).
Approximately 100 seeds (2.5 g) were suspended in 50 mL TSB of each bacterial suspension. In the co-inoculation treatment the culture suspensions of Az39 and G16 strains were mixed immediately before the seeds were soaked into the mixed bacterial suspension. Non-inoculated seeds were suspended in sterile TSB. The seeds were shaken for 100 min to allow the penetration of bacteria into the seeds. The medium was removed and seeds were transferred to sterilized wet gauze pads in Petri dishes and incubated in the dark for 5 days at 30ºC for germination.
Plants growth and yield
Two germinated seeds were transferred aseptically to a 100 mL culture tube containing sterilized expanded perlite (Perliv ® ) and 30 mL of 1:2 diluted Yoshida medium (modified from Gregorio et al., 1997). Hydroponic cultivation of plantlets was performed under gnotobiotic conditions for 10 days. After incubation, between 13 and 19 seedlings of each block were harvested, pooled and analysed. The dry weight of the entire plant was determined. Additionally, the fresh weight and length of the roots and aerial portions were measured.
Counts of endophytic, heterotrophic, siderophore-producing and diazotrophic bacteria
The roots and shoots of four seedlings from each replicate were analysed separately. Two hundred milligrams of each plant tissue was surface-sterilized by shaking for 5 min in 100 mL of NaClO 1.7 %. Four washes in 200 mL of sterilized distilled water were used to remove the disinfectant.
Each plant tissue sample was transferred to a mortar and homogenized in 4 mL of saline solution. The suspensions were serially diluted with sterile 0.9% NaCl and plated onto R2A for total heterotrophic bacteria (TH) counts or in R2A-CAS media (Schwyn and Neilands, 1987) modified by reducing the PIPES final concentration to 50 mM for siderophore-producing heterotrophic bacteria (SPH) counts. Plates were incubated at 30ºC for 72 h. Counts were expressed as the number of colony-forming units (CFU) per gram of fresh weight. The average number of bacteria among duplicated blocks and the standard deviation were calculated.
The density of endophytic diazotrophic bacteria was determined using the most probable number (MPN) counts in RMR medium (Elbeltagy et al., 2001) without vegetal extract. Culture tubes showing turbidity and acetylene reduction (Loaces et al., 2011) were considered positive for diazotrophs.
Isolation, identification, and physiological charac-terization of endophytic bacteria
Endophytes from seedlings. To identify the endophytes recovered from inoculated and non-inoculated seedlings, bacteria obtained from plate counts on different media were isolated. Colonies with different morphology from the higher dilution counts in CAS-R2A and R2A media were purified and identified by 16S rRNA gene sequencing as described below.
Several seed-borne endophytes were further characterized. The isolates were tested for their ability to fix nitrogen, solubilize phosphate, and produce siderophores and indol acetic acid (IAA). Nitrogen fixation was evaluated by the acetylene reduction assay in 27 mL vials containing 18 mL of RMR semisolid medium. After incubation at 30ºC for 7 days in the dark, acetylene gas was added to the headspace atmosphere of the vials at a final concentration of 10% (v/v) and incubated at 30ºC. After 5 days of incubation, ethylene was determined by gas chromatography as described by Loaces et al., (2011). A. brasilense Az39 was used as a positive control, and non-inoculated tubes were used as negative controls. Inorganic phosphate solubilization was confirmed using Pikovskaya agar (Pikovskaya, 1948) with cycloheximide (50 mg/l) after incubation at 30°C for 7 days. Siderophore-producing activity was detected on R2A-CAS medium by the orange-yellow halo around the colony. Indole acetic acid production was determined in medium containing 20 g/l of bacteriological peptone (Oxoid, Basingstoke, UK), 1.15 g/l K2HPO4, 1.5 g/l MgSO4.7H2O and 2.5 mM of tryptophan (Merck, Darmstadt, Germany). After 12 h of incubation, 1 ml of supernatant was mixed with 1 ml of Salkowski reagent and analysed as it was described in a previous work (Loaces et al., 2011). The analyses were performed in duplicate.
Endophytes from seeds
To examine the presence of P. oryzihabitans in other rice varieties relevant to Uruguay, 5 g of seeds of Oryza sativa varieties INIA-Tacuarí and El Paso 144 were peeled and surface-sterilized as described above. After maceration using a mortar, saline ten-fold dilutions were prepared and plated in R2A and R2A-CAS. Plates were incubated at 30ºC for 5 days. Colonies with a similar morphology to P. oryzihabitans G16 were purified, and DNA was extracted for further identification using 16S rRNA gene sequencing as described below. The isolates were tested for antagonistic activity against A. brasilense Az39 as it was described in a previous work (Loaces et al., 2011).
Molecular characterization of the isolates
16S rRNA gene amplification and sequencing. The isolates were screened and identified by 16S rRNA gene analysis (Loaces et al., 2011). The colonies were suspended in sterilized water, and the suspension was centrifuged (10 min at 15,000×g, 4ºC). DNA was extracted from the pellets using the Wizard Genomic DNA Purification Kit (Promega).
16S rDNA amplification was performed in 25 μL of reaction mixture that contained: 0.48 μM of primers (IDT, USA) 27f (5-AGAGTTTGATCCTGGCTCAG-3´) and 1492r (5'-GGTTACCTTGTTACGACTT-3´), 0.2 mM of each dNTP (Fermentas), 2.5 mM MgCl2, 0.25 mg/ml of bovine serum albumin (Roche), 2.5 μL of Taq buffer and 1 U of Taq DNA polymerase (Fermentas). PCR amplification was performed on 1 μL of DNA template in an automated thermal cycler (2720 Thermal Cycler, Applied Biosystems) with an initial denaturation (94ºC for 5 min) followed by 30 cycles of amplification (94ºC for 1 min, 55ºC for 1 min, and 72ºC for 3 min) and a single final extension (72ºC for 7 min).
16S rRNA gene amplicons were screened by ARDRA (Amplified Ribosomal DNA Restriction Analysis) to select representative ribotypes of all the isolates. PCR products were digested with 3 units each of MspI (Fermentas) and RsaI (Fermentas) at 37ºC for 10 h. The restriction fragments were separated on a 3.5 % agarose (Methaphor, FMC, USA) gel running in 0.5 × TBE buffer for 1 h at 100 mV and stained with ethidium bromide. A computer-assisted evaluation of the ARDRA patterns generated was made using the GelCompar 4.2 program (Applied Maths, Kortrijk, Belgium). Restricted amplicons were grouped into operational taxonomic units (OTUs) based on these restriction patterns.
Representative ribotypes according to the ARDRA profile were selected, and the 16S rRNA genes were partially sequenced using the primer sets described above. All sequencing reactions were performed by the Macrogen Sequencing Service, Korea using an ABI PRISM 3730XL capillary sequencer (Applied Biosystems, Foster City, USA). The sequences were submitted to the National Center for Biotechnology Information database, and the closest relative sequence was used for identification.
The sequences of the isolates obtained from seeds of the varieties INIA-Tacuarí and El Paso 144 are available at the GenBank database with the accession numbers: KM225652 to KM225657.
nifH gene amplification. All of the isolated strains were tested for the presence of the nifH gene. Primers previously described (Zehr and McReynolds, 1989; Poly et al., 2001) were used for screening. The nifH gene amplification and PCR conditions for the primer set designed by Poly et al. (2001) were performed according to a previous work (Ferrando and Fernández Scavino, 2015). PCR reactions using the primer set from Zher and McReynolds (1989) (nifHZMRf, 5’-TGYGAYCNNAARGCNGA-3´; nifHZMRr, 5’-ADNGCCATCATYTCNCC-3´) were performed in 25 µL (total volume) mixtures containing approx. 100- 200 ng of total DNA, 0.1 mmol/L of each primer, 2.5 mmol/L MgCl2, Taq buffer, Bovine Serum Albumin 0.2 mg/mL (Roche®), 0.2 mmol/L of each dNTP and 1.2 U of Taq DNA polymerase (Fermentas ©). The reactions were performed in an Applied Biosystems 2720 Thermal Cycler (Singapur) using the following program: initial denaturation step at 94ºC for 5 min, followed by 30 cycles at 94ºC for 60 s, 57ºC for 60 s and 72ºC for 60 s, with a final extension step at 72ºC for 10 min. The amplicons obtained were confirmed by electrophoresis on 1 % agarose gel (Agarose I, Amresco) at 100 V.
Statistical analyses
Statistical analyses were performed using InfoStat (Di Renzo et al., 2009). Plate counts (HB and HSPB) and vegetal yield data were subjected to analysis of variance (ANOVA). Differences were considered significant at the 95% (of higher) confidence level. Data related to the CFU were transformed into logarithmic values before statistical analysis.
Results
Effect of inoculation on plant yield
Plant growth was affected by the inoculated bacteria. After 15 days of seed inoculation, shoot fresh weight was significantly higher in seedlings inoculated with strain Az39 than in seedlings inoculated with strain G16 (Table 1). Additionally, the same trend was observed for most of the parameters measured, with Az39 stimulating growth and G16-inoculated seedlings showing the lowest yield values. Non-inoculated or co-inoculated seedlings showed intermediate values for all of the parameters measured.
Bacterial abundance in non-inoculated and in-oculated plants
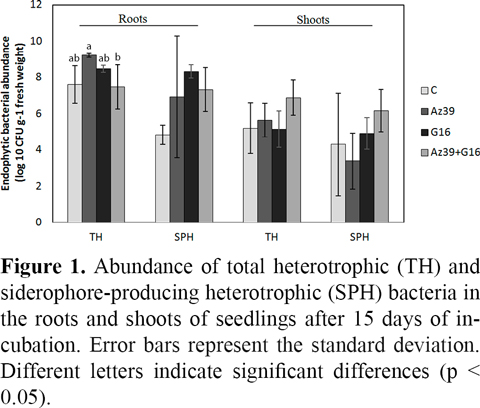
Total heterotrophic (TH) and siderophore-producing heterotrophic (SPH) bacteria were counted in the shoots and roots of seedlings after 15 days of incubation to compare the population density of endophytic bacteria in different inoculation treatments.
The abundance of the total heterotrophic bacteria expressed as the log CFU/g fresh tissue was between 5.2 and 6.9 in shoots and between 7.5 and 9.2 in roots of all seedlings. Non-inoculated plants exhibited a high density of heterotrophic bacteria with counts that were not significantly different from the co-inoculated or G16-inoculated plants. These results indicate that seed-borne bacteria were able to grow in the roots and shoots of rice, reaching similar densities as those of inoculated plants.
Inoculation with Azospirillum significantly increased the heterotrophic bacteria in roots compared with non-inoculated plants (Figure 1), suggesting that Az39 could grow or increase the population size of seed-borne endophytes in roots. Strain G16 also increased the bacterial counts, although the values were not significantly different from other treatments. On the other hand, co-inoculated and non-inoculated seedlings showed a similar density of heterotrophic bacteria, indicating that mixed inoculation did not produce a significant increase of heterotrophic endophytes.
R2A-CAS medium was used to improve the recovery of the inoculated strains, especially because non-siderophore producing seed-borne endophytes were abundant, even in inoculated plants (Fig. 1). Siderophore producing bacteria were in the range of 3.4 - 6.2 log CFU/g in shoots and 4.8 - 8.3 log CFU/g in roots. SPB were the lowest in non-inoculated plants, although no significant differences were observed among treatments in the roots or shoots. Since strains Az39 and G16 produce siderophores, the high density of SPB observed in the inoculated plants was not surprising.
Diazotrophic bacteria were below the detection limit of the method (60 MPN diazotrophs/g fresh weight) for non-inoculated and G16-inoculated plants. These results indicate that most of the seed-borne endophytes were not able to fix N2. In Az39-inoculated seedlings, the abundance of diazotrophs (MPN/g fresh weight) was 4.6 x 102 in the shoots and 4.8 x 104 in the roots for both replicates. In co-inoculated plants, diazotrophs were only recovered from the shoots of one block (8.0 x 101/g fresh weight), suggesting that inoculation with strain G16 decreased the density of Az39.
Identification of endophytic bacteria
Thebacteria recovered from R2A and R2A-CAS count plates were purified and sixty isolates were selected for molecular characterization. The genetic profiles of these isolates were screened by ARDRA, and eight different OTUs were obtained. Several isolates representative of each OTU profile were identified by 16S rRNA gene partial sequencing.
Endophytic bacteria from inoculated seedlings
The predominant bacteria recovered from inoculated plants were the bacteria inoculated, except in co-inoculated seedlings. Table 2 shows the density of the inoculated bacteria, when recovered, in inoculated plants. In single-inoculated seedlings A. brasilense or P. oryzihabitans were the most abundant bacteria recovered. However, in co-inoculated plants, A. brasilense was not recovered from R2A or R2A-CAS count plates and P. oryzihabitans was the most abundant heterotrophic bacteria detected in both vegetal tissues.
Seed-borne endophytic bacteria
The dominant species recovered in both replicates from uninoculated seedlings was Sphingomonas yabuuchiae. In these plants, the species reached counts of 7.5 ± 1.1 log CFU/g fresh weight of roots and 5.2 ± 1.4 log CFU/g fresh weight of shoots. Furthermore, this species was present in shoots and roots of all inoculated plants (Table 3), indicating that this seed-borne endophyte was not excluded by competition by P. oryzihabitans or A. brasilense.
The seed-borne endophytes isolated from inoculated seedlings were in lower densities than A. brasilense or P. oryzihabitans. Members of the genus Pantoea were less ubiquitous than S.yabuuchiae, being detected in non-inoculated (OTU B) or P. oryzihabitans-inoculated (OTU J) seedlings (Table 3). Bacteria from the genera Methylobacterium, Microbacterium and Chryseobacterium were also isolated, but usually they were restricted to one treatment or to one block of one treatment.
Pseudomonas oryzihabitans (strain ED1) was recovered from the roots of uninoculated plants in one of two replicated blocks with a density three orders lower than the predominant S. yabuuchiae.
Physiological characterization of seed-borne endophytic bacteria
Four strains of seed-borne endophytic bacteria were further characterized regarding their physiological traits relevant for plant promoting growth or ability to outcompete A.brasilense or P.oryzihabitans. None were able to fix N2, and the nifH gene was not detected. However, most of the isolates exhibited other valuable properties, such as phosphate solubilization, siderophore production and IAA production (Table 4). Interestingly, the highest IAA producer was the ubiquitous endophyte: Sphingomonas yabuuchiae.
Detection of P. oryzihabitans in seeds of different rice varieties
Seeds of O. sativa var. INIA-Tacuarí and El Paso 144 were screened for bacteria morphologically similar to P. oryzihabitans G16. The isolates obtained in R2A-CAS plates were purified and identified by 16S rRNA gene partial sequencing (Table 5).
Several siderophore-producing bacteria similar to P. oryzihabitans G16 were isolated from seeds of both varieties. These strains could be identified as P. oryzihabitans, very closely related to the type strain (NBRC 102199). All of them, except strain 1B, were able to inhibit A. brasilense Az39 in agar plates.
Discussion and conclussions
The development of inoculant technology is a valuable alternative to improve crop productivity and sustainability of agriculture since inoculants reduce the utilization of chemicals, minimizing pollution and the consumption of non-renewable energy sources. Moreover, the use of native PGPB contributes to the valorization of regional genetic resources and enhances the probability of adaptation of the local biota, especially to address the stress conditions imposed by climate change.
The challenges to expanding this technology lie in the better understanding of the influences of the biotic and abiotic parameters on the biological interactions involved. The success of the inoculation is mainly attributed to the early fitting of PGPB to the plant. However, this is not a simple two-partner relationship since a complex community of microorganisms from the seeds and soil influence the colonization and activity of the PGPB. Thus, the agronomic potential of PGPB depends on the density and activity of a diverse group of microorganisms that can be synergic, neutral or antagonistic to the inoculated bacteria.
In this work, a gnotobiotic experiment was set up to determine the effect of the inoculation of similar amounts of a seed-borne rice endophytic bacteria, P. oryzihabitans, on A. brasilense-inoculated seedlings. The effects of the inoculation or co-inoculation on plant yield and on the native endophytic bacterial community were also determined.
The impact of the inoculation on plant growth was studied for the whole plant, but significant differences were observed only in the shoot biomass. A. brasilense Az39 had a growth promotion effect, whereas P. oryzihabitans G16 was deleterious for plant development. Co-inoculated seedlings exhibited similar vegetal yield values as non-inoculated seedlings, suggesting that the opposite effects caused by strains G16 and Az39 were neutralized when both strains were inoculated at high densities. The ability of A. brasilense Az39 to promote vegetal growth has been well documented in field trials (Fulchieri and Frioni, 1994; Cassán et al., 2009; Zawoznik et al., 2011; Masciarelli et al., 2013; Díaz-Zorita et al., 2015; Pereg et al., 2016), but the harmful effect of members of the species P. oryzihabitans on plant growth has not been reported. Although bacteria closely related to this species are common endophytes of rice seeds and plants (Cottyn et al., 2009; Ruiza et al., 2011; Hardoim et al., 2012; Hameed et al., 2015), the harmful effect may only be observed when this microorganism is a prominent member of the endophytic community.
Non-inoculated and inoculated seedlings harboured a high density of heterotrophic and siderophore-producing bacteria in the roots and shoots (Figure 1). The high density of endophytes in the control plants indicates that the seed-borne endophytes were able to grow in planta, reaching similar densities as in the inoculated plants. Moreover, these results showed that, irrespective of the inoculation, endophytes in seedlings incubated under gnotobiotic conditions can be as high as in seedlings cultivated in soil (Mano et al., 2007; Ferrando et al., 2012; Hameed et al., 2015).
Inoculation with A. brasilense Az39 significantly increased heterotrophic counts compared with non-inoculated or co-inoculated roots (Figure 1). A. brasilense was the predominant species identified in these seedlings (Table 2), suggesting that strain Az39 was able to overgrow the population of seed-borne endophytes in the roots and shoots. Similarly, P.oryzihabitans was predominant in the shoots and roots of G16-inoculated plants. These results confirm that both species are endophytes capable of root and shoot colonization and, when inoculated in high amounts, they can prevail over the seed-borne endophytes.
Unexpectedly, only P. oryzihabitans was retrieved as the predominant species in the shoots and roots of co-inoculated plants (Table 2). In agreement with these results, whereas diazotrophs were quantified in the shoots and roots of Az39 inoculated plants, only the shoots of plants from one replicate was positive for diazotrophs in co-inoculated plants. Since P. oryzihabitans G16 does not have the ability to fix nitrogen and the seed-borne diazotrophs were below the quantification limit, positive tubes in diazotrophs counts can be attributed to the presence of A. brasilense. These results indicate that P. oryzihabitans G16 overgrew A. brasilense Az39 in plant tissues when the seeds of rice were co-inoculated with similar amounts of both strains. Antagonism of P. oryzihabitans G16 towards PGPB has been previously observed in agar plates (Loaces et al., 2011), but this activity has not been proved in planta. Noticeably, there are many commercial inoculants with combined formulations composed of A. brasilense and biocontrol agents belonging to the genus Pseudomonas, particularly P. fluorescens (Valverde et al., 2015). The combination of phytobeneficial microorganisms with different metabolic capacities is frequent in commercial inoculants, but their mutual exclusion has been scarcely tested. This is particularly relevant since many mechanisms of biocontrol towards plant pathogens could be the same as those against beneficial bacteria. It has been shown that Pseudomonas fluorescens decreased the abundance of A. brasilense and its phytostimulation capability in wheat roots (Couillerot et al., 2011). These authors found that the antimicrobial compound 2,4-diacetylphloroglucinol (DAPG) produced by Pseudomonas is responsible for such inhibition. It was also observed that different species of Azospirillum can be resistant to DAPG, although all of them decayed in the rhizosphere of maize when they were co-inoculated with P. fluorescens F113 (Couillerot et al., 2013). Thus, the interaction between Azospirillum spp. and P. fluorescens could be complex and require more than one compound.
Although P. oryzihabitans G16 was isolated from seeds of the variety INIA-Olimar (type Indica) of O. sativa, strains of this species were isolated from seeds of a variety of the same type (El Paso 144) or of the type Japonica (INIA-Tacuarí) (Table 5). The six strains identified in this work produced siderophores, and five of them were able to antagonize A. brasilense Az39 in an agar plate assay. Additionally, P. oryzihabitans was detected in non-inoculated seedlings incubated under hydroponic conditions (strain ED1, Table 3), indicating that this microorganism can grow in plant tissues and can be translocated to new seeds and thus be preserved as endophyte with the reproductive cycle of the rice. It was postulated that seeds are a source of bacteria that can be dispersed into rhizospheric soil (Hardoim et al., 2012). Our results showed that bacteria closely related to strain P. oryzihabitans G16 were widespread among rice varieties cultivated in Uruguay, kept in seeds and probably dispersed by rice-rhizospheric soil.
Antagonism between A. brasilense and seed-borne Gram positive bacteria in rice rhizoplane of 15 days-old plantlets has been observed by Bacilio-Jiménez et al. (2001), although the mechanism involved has not been explored. These authors observed that the root surface was colonized preferentially by endophytes from the genera Bacillus and Corynebacterium, which promoted the growth of roots, whereas A.brasilense inhibited the growth of rice. In our case, strains of P. oryzihabitans that antagonize A. brasilense in vitro showed a high ability to capture Fe+3. Thus, as it has been reported for many bacteria, siderophore production could be a plausible mechanism to enforce biocontrol capabilities (Miethke and Marahiel, 2007; Fernández and Pedraza, 2013). Furthermore, we observed that the iron-mediated competence of strain G16 did not suppress the growth of endophytes with a noticeable ability to produce siderophores (Table 3 and Table 4). Members of the genus Sphingomonas, prominent in non-inoculated plants, were found in all treatments, suggesting that these bacteria have the highest fitness to O. sativa during the early steps of plant growth, even when high amounts of other inoculated bacteria were present. Bacteria from the genus Sphingomonas have been reported previously as endophytes of rice (Ferrando et al., 2012; Zhang et al., 2013). Pantoea sp. were also present in uninoculated plants and able to thrive in plants inoculated with strain G16 (Table 3). This genus has been frequently found among the culturable endophytic bacteria in rice (Verma et al., 2004; Mano et al., 2006; Prakamhang et al., 2009; Loaces et al., 2011). Interestingly, bacteria from both genera also showed traits involved in vegetal growth promotion, such as the capability to solubilize inorganic phosphate or to produce IAA (Table 4). Therefore, siderophore-producing seed-borne bacteria with phytostimulatory properties were not excluded from plant tissues by inoculation with hign amounts of P. oryzihabitans G16.
In summary, the present work shows that high amounts of the strain G16 of P. oryzihabitans, an endophytic species common in rice seeds, can damage young plants when present at high densities. In these conditions, P. oryzihabitans outcompetes A. brasilense Az39, the most commonly used strain in commercial inoculants for cereals in South America. On the other hand, the antagonistic capability of P. oryzihabitans seems to be selective, since other seed-borne endophytes, such as Sphingomonas sp. or Pantoea sp., which are strong siderophore producers, were not inhibited in planta. Moreover, high amounts of A. brasilense Az39 stimulated the growth of young plants and prevailed over the harmful P. oryzihabitans present in low amounts in rice seeds. Therefore, the stimulation of A. brasilense, commonly attributed to phytohormone production, could also, or alternatively, be due to the modulation of the native, potentially detrimental, bacteria associated with the plant.
Acknowledgments
We thank Guilermo Arrospide from Calister S.A. for providing the strain A. brasilense Az39, and Enrique Deambrosi and José Terra from INIA Treinta y Tres for providing seeds of different rice varieties. Thanks to Silvana Vero and Inés Loaces for technical assistance. This work was partially supported by PEDECIBA (Programa de Desarrollo de la Ciencias Básicas) and CSIC (Comisión Sectorial de Investigación Científica).
References
1. Araújo A.E., da S., Baldani V.L.D., Galisa P. de S., Pereira J.A., Baldani J.I. (2013). Response of traditional upland rice varieties to inoculation with selected diazotrophic bacteria isolated from rice cropped at the Northeast region of Brazil. Applied Soil Ecology 64: 49-55. [ Links ]
2. Bacilio-Jimenez M., Aguilar-Flores S., del Valle M., Perez A., Zepeda A., Zenteno E. (2001). Endophytic bacteria in rice seeds inhibit early colonization of roots by Azospirillum brasilense. Soil Biology Biochemistry 33 (2): 167-172. [ Links ]
3. Bashan Y., de-Bashan L. (2010). How the plant growth-promoting bacterium Azospirillum promotes plant growth a critical assessment. Advances in Agronomy 108 (2): 77–136.
4. Cassán F., Díaz-Zorita M. (2016). Azospirillum sp. in current agriculture: From the laboratory to the field. Soil Biology Biochemistry 103: 117-130. [ Links ]
5. Cassán F., Maiale S., Masciarelli O., Vidal A., Luna V., Ruiz O. (2009). Cadaverine production by Azospirillum brasilense and its possible role in plant growth promotion and osmotic stress mitigation. European Journal of Soil Biology 45 (1): 12-19. [ Links ]
6. Cottyn B., Debode J., Regalado E., Mew T.W., Swings J. (2009). Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. Journal of Applied Microbiology 107 (3): 885-897. [ Links ]
7. Couillerot O., Combes-Meynet E., Pothier J.F., Bellvert F., Challita E., Poirier M.A., Rohr R., Comte G., Moënne-Loccoz Y., Prigent-Combaret Y. (2011). The role of the antimicrobial compound 2,4-diacetylphloroglucinol in the impact of biocontrol Pseudomonas fluorescens F113 on Azospirillum brasilense phytostimulators. Microbiology 157 (6): 1694-1705. [ Links ]
8. Couillerot O., Ramirez-Trujillo A., Walker V., von Felten A., Jansa J., Maurhofer M., Defago G., Prigent-Combaret C., Comte G., Caballero-Mellado J., Moenne-Loccoz Y. (2013). Comparison of prominent Azospirillum strains in Azospirillum-Pseudomonas-Glomus consortia for promotion of maize growth. Applied Microbiology and Biotechnology 97 (10): 4639-4649. [ Links ]
9. Di Rienzo J.A., Casanoves F., Balzarini M.G., Gonzalez L., Tablada M., Robledo C.W. (2009). Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. [ Links ]
10. Díaz-Zorita M., Canigia M.V.F., Bravo O.Á., Berger A., Satorre E.H. (2015). Field Evaluation of Extensive Crops Inoculated with Azospirillum sp. In: Handbook for Azospirillum. Cassán F.D., Okon Y., Creus C.M. (Eds.). Springer International Publishing Switzerland. Pp. 435-445. [ Links ]
11. Elbeltagy A., Nishioka K., Sato T., Suzuki H., Ye B., Hamada T., Isawa T., Mitsui H., Minamisawa K. (2001). Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Applied Environmental Microbiology 67 (11): 5285-5293. [ Links ]
12. Fernández S.A., Pedraza R.O. (2013). The role of siderophores in plant growth-promoting bacteria. In: Bacteria in Agrobiology: Crop Productivity. Maheshwari D.K., Saraf M., Aeron A. (Eds.). Springer-Verlag, Germany. Pp. 265-285. [ Links ]
13. Ferrando L., Fernández Scavino A. (2015). Strong shift in the diazotrophic endophytic bacterial community inhabiting rice (Oryza sativa) plants after flooding. FEMS Microbiology Ecology doi: 10.1093/femsec/fiv104. [ Links ]
14. Ferrando L., Mañay J.F., Scavino A.F. (2012). Molecular and culture-dependent analyses revealed similarities in the endophytic bacterial community composition of leaves from three rice (Oryza sativa) varieties. FEMS Microbiology Ecology 80 (3): 696-708. [ Links ]
15. Fulchieri M., Frioni L. (1994). Azospirillum inoculation on maize (Zea mays L.): effects on yield in a field experiment in central Argentina. Soil Biology Biochemistry 26: 921-923. [ Links ]
16. García de Salamone I.E., Funes J.M., Di Salvo L.P., Escobar-Ortega J.S., D’Auria F., Ferrando L., Fernandez-Scavino A. (2012). Inoculation of paddy rice with Azospirillum brasilense and Pseudomonas fluorescens: impact of plant genotypes on rhizosphere microbial communities and field crop production. Applied Soil Ecology 61: 196-204.
17. Gregorio G.B., Senadhira D., Mendoza R.D. (1997). Screening rice for salinity tolerance. Plant Breedin, Genetics, and Biochemistry Division. IRRI discussion paper series 22: 1-30. [ Links ]
18. Hallmann J., Quadt-Hallmann A., Mahaffee W.F., Kloepper J.W. (1997). Bacterial endophytes in agricultural crops. Canadian Journal Microbiology 43 (10): 895-914. [ Links ]
19. Hameed A., Yeh M.W., Hsieh Y.T., Chung W.C., Lo C.T., Young L.S. (2015). Diversity and functional characterization of bacterial endophytes dwelling in various rice (Oryza sativa L.) tissues, and their seed borne dissemination into rhizosphere under gnotobiotic P- stress. Plant Soil 394 (1): 177-197. [ Links ]
20. Hardoim P.R., Hardoim C.C., Van Overbeek L.S., Van Elsas J.D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One. doi:10.1371/journal.pone.0030438. [ Links ]
21. Hungria M., Nogueira M., Araujo R. (2013). Co-inoculation of soybeans and common beans with rhizobia and azospirilla: strategies to improve sustainability. Biology Fertility of Soils 49 (7): 791-801. [ Links ]
22. Loaces I., Ferrando L., Scavino A.F. (2011). Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microbiology Ecology 61 (3): 606-618. [ Links ]
23. Mano H., Tanaka F., Nakamura C., Kaga H., Morisaki H. (2007). Culturable Endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) cultivated in a paddy soil. Microbes Environment 22 (2): 175-185. [ Links ]
24. Mano H., Tanaka F., Nakamura C., Watanabe A., Kaga H., Okunishi S., Morisaki H. (2006). Culturable surface and endophytic bacterial flora of the maturing seeds of rice plants (Oryza sativa) cultivated in a paddy field. Microbes Environment 21 (2): 86-100. [ Links ]
25. Masciarelli O., Urbani L., Reinoso H., Luna V. (2013). Alternative mechanism for the evaluation of indole-3-acetic acid (IAA) production by Azospirillum brasilense strains and its effects on the germination and growth of maize seedlings. Journal of Microbiology 51 (5): 590-597. [ Links ]
26. Miethke M., Marahiel M. (2007). Siderophore-based iron acquisition and pathogen control. Microbiology and Molecular Biology Reviews 71 (3): 413-451. [ Links ]
27. Muñoz-Rojas J., Fuentes-Ramírez J.E., Caballero-Mellado J. (2005). Antagonism among Gluconacetobacter diazotrophicus strains in culture media and in endophytic association. FEMS Microbiology Ecology 54: 57-66. [ Links ]
28. Okunishi S., Sako K., Mano H., Imamura A., Morisaki H. (2005). Bacterial flora of endophytes in the maturing seed of cultivated rice (Oryza sativa). Microbes Environment 20 (3): 168-177. [ Links ]
29. Pereg L., de-Bashan L.E., Bashan Y. (2016). Assessment of affinity and specificity of Azospirillum for plants. Plant Soil 399 (1): 389-414. [ Links ]
30. Piccinin G., Dan L., Braccini A., Mariano D., Okumura R., Bazo G., Ricci T. (2011). Agronomic efficiency of Azospirillum brasilense in physiological parameters and yield components in wheat crop. Journal of Agronomy 10 (4): 132-135. [ Links ]
31. Pikovskaya R.I. (1948). Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Mikrobiologiya 17:362-370. [ Links ]
32. Poly F., Jocteur Monrozier L., Bally R. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Research Microbiology 152 (1): 95-103. [ Links ]
33. Prakamhang J., Minamisawa K., Teamtaisong K., Boonkerd N., Teaumroong N. (2009). The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L.). Applied Soil Ecology 42 (2): 141-149. [ Links ]
34. Ruiza D., Agaras B., de Werra P., Wall L.G., Valverde C. (2011). Characterization and screening of plant probiotic traits of bacteria isolated from rice seeds cultivated in Argentina. Journal of Microbiology 49 (6): 902-912. [ Links ]
35. Schwyn B., Neilands J.B. (1987). Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry 160 (1): 47-56. [ Links ]
36. Valverde C., Anta G.G., Ferraris G. (2015). Pseudomonas and Azospirillum. In: Handbook for Azospirillum. Cassán F.D., Okon Y., Creus C.M. (Eds.) Springer International Publishing, Switzerland. Pp. 389-409. [ Links ]
37. Verma S.C., Singh A., Chowdhury S.P., Tripathi A.K. (2004). Endophytic colonization ability of two deep-water rice endophytes, Pantoea sp. and Ochrobactrum sp. using green fluorescent protein reporter. Biotechnology Letters 26 (5): 425-429. [ Links ]
38. Zawoznik M.S., Ameneiros M., Benavides M.P., Vázquez S., Groppa M.D. (2011). Response to saline stress and aquaporin expression in Azospirillum-inoculated barley seedlings. Applied Microbiology Biotechnology 90 (4): 1389-1397. [ Links ]
39. Zehr J.P., McReynolds L.A. (1989). The use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Applied Environmental Microbiology 55 (10): 2522-2526. [ Links ]
40. Zhang X.X., Gao J.S., Cao Y.H., Ma X.T., He J.Z. (2013). Long-Term rice and green manure rotation alters the endophytic bacterial communities of the rice root. Microbiology Ecology 66: 917-926. [ Links ]