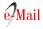Introduction
Argentina ranks sixth in maize production, with approximately 60 million tons a year. The region encompassing Santiago del Estero province, east of Tucumán and north of Córdoba, in the Chaco Semi-arid region, produces about 6 million tons (58). This geographical area is a critical climatic zone, where climatic factors are highly variable (57). Thus, maize crop development in these agroecosystems is constrained by stressful environmental factors that expose the crop to different pathogens, such as those responsible for ear rot (27,67). The most dangerous of these pathogens is Aspergillus flavus Link, a globally distributed filamentous, cosmopolitan pathogen that causes opportunistic infections in animals and plants (12,42). This pathogen is commonly present in air and soil mycobiota. Under certain water and thermal stress conditions, some isolates produce mycotoxins like aflatoxins and cyclopiazonic acid (CPA). Aflatoxins are secondary metabolites with strong hepatotoxic, teratogenic, immunosuppressive and mutagenic activity when inhaled, ingested or absorbed through animal and human skin (51); A. flavus is the main driver of aflatoxin contamination in maize in the world (45). While CPA produces liver necrosis, convulsions myocardium lesions in animals (62,63), the specific exposure response to this toxin differs among species, without sufficient evidence about natural contamination of foods (81).
The maximum safety limit of aflatoxins for maize commercialization varies between countries, being 20 ng. g-1 in Argentina, Brazil and USA, 4 ng. g-1 in the European Union, and 15 ng. g-1 in African countries like Ghana (4,9,16,34,77).
Species of the genus Aspergillus produce sclerotia, resistance structures consisting of darkly pigmented and hardened hyphal mats (50) conferring different phenotypic characteristics (1). Thus, Aspergillus species can be classified into two morphotypes: S isolates, which produce numerous sclerotia under 400 μm in diameter, associated with high levels of aflatoxins, and L isolates, with fewer sclerotia, diameter over 400 μm, and related to variable levels of aflatoxins (75). A third type of isolate not producing sclerotia under laboratory conditions, has been identified in Argentina and Italy (13,38).
The variability among A. flavus populations can be estimated by establishing phylogenetic relationships based on morphological and molecular markers (51), as in the case of the calmodulin gene (CaM) (36). Calmodulin (CaM) is a highly conserved polypeptide present in eukaryotic cells. In the genus Aspergillus, CaM is important in phosphorylation and dephosphorylation of proteins involved in the biosynthesis of aflatoxins (47).
Aflatoxin contamination usually starts in the field, during harvest of infected grains, becoming more severe during storage (35). In the field, spores of A. flavus are transported by biotic agents (insects, birds) and/or abiotic agents (wind, rain splash) to the maize ear. Spores enter through the silk during flowering and through the wounds of the ear cover or of developing kernels (66). Water stress, high temperatures, insect attack, certain sowing dates and high crop density favor contamination with the pathogen and the possible production of aflatoxins (48).
Some members of A. flavus were found unable to biosynthesize aflatoxins being, therefore, plausible biocontrol tools (23). Biological control applied in maize and peanut (6,73), among other crops, is based on the competition for infection sites and essential nutrients between non-aflatoxigenic and aflatoxigenic isolates. Thus, non-aflatoxigenic isolates displace aflatoxigenic ones and reduce the latter’s capacity to produce aflatoxins (70,79).
Non-aflatoxin producing strains selected as biocontrol agents must meet some requirements, such as being native to the area of application, being able to displace the toxicogenic isolates in the ecosystem and thereby reduce infections (23) and be genetically incompatible with the remaining fungal population. Therefore, studies of the native fungal population are crucial (42).
This work aimed to study the diversity of A. flavus populations in the Chaco Semi-arid region of Argentina through the morphological, toxicological and molecular characterization of isolates obtained from maize ears.
Materials and methods
Sampling
Sampling was conducted during the 2015/16 and 2016/17 growing seasons in representative localities of eight agro-climatic zones: Sachayoj (Zone 4: Río Muerto), Quimilí (Zone 5: Hickmann), Bandera (Zone 17: Bandera), Santiago del Estero (Zone 18: Monteagudo), Sumampa (Zone 37: Soto), Santa Rosa de Leales (Zone 18A: Monteagudo), Rayo Cortado (Zone 37C: Soto-north of Córdoba) and Villa de Tulumba (Zone 61B: Deán Funes), located between -30.38 and -26.55 S and -61.82 and -65.27 W (25) (figure 1, page 61).
Sampling localities / Localidades evaluadas: Sachayoj. Quimilí. Santiago del Estero. Bandera. Sumampa (Santiago del Estero province). Santa Rosa de Leales (Tucumán province). Rayo Cortado. Villa de Tulumba (Córdoba province).
Figure 1 Sampling localities in the Chaco Semi-arid region of Argentina. Figura 1. Localidades evaluadas en la región del Chaco semiárido argentino.
Ten ears were randomly collected along a diagonal line in the plot, at plant physiological maturity before harvest. They were threshed to form a composite sample and oven-dried at 38°C for 72 hours until final humidity was under 12 %, therefore, minimizing fungus development (26).
Isolation and morphological and molecular identification
From each sample, 100 kernels were taken, surface-desinfested with a 1% sodium hypochlorite wash followed by three washes with distilled water and plated in DG-18 medium. Kernels were incubated for 7 days at 25°C and cultures with characteristics similar to those of A. flavus were transferred to MEA medium. They were incubated for 7 days at 25°C (65) and then identified through taxonomic keys (48). Spores of each isolate were serially diluted, and the most diluted concentration was cultured in 2 % Water - Agar (WA) medium and incubated for 18 hours at 27°C (65). The germinated conidia were identified under Nikon SMZ-10 stereoscopic microscope (15X). Two conidia were extracted from each cultured Petri dish, along with a portion of culture medium, and transferred to Petri dishes containing MEA medium. They were incubated for 7 days at 25°C. Then, one of them was stored to obtain the single spore cultures corresponding to each isolate.
The identity of A. flavus was confirmed by PCR using the primers FLA1, 5’-GTAGGGTTCCTAGCGAGCC-3’; FLA2 5’-GGAAAAAGATTGATTTGCGTTC-3’ (41), with the ITS region as target, using the isolate A. flavus CCC 101-92 < NRRL 3251 as positive control, and the isolate A. parasiticus NRRL 2999 as negative control. Each single-spore isolate was cultured in MEA and incubated for 7 days at 25°C. Then, an aliquot of mycelium was taken, and DNA was extracted (55). For each reaction, 1 μL (final concentration of 20-100 ng of DNA.μL-1, quantified in Nano Drop, Thermo Fisher Scientific, USA) of DNA solution was mixed with 24 μL of a solution composed of 5 μL 5x Green GoTaq® reaction buffer (Promega®, Madison, WI, USA), 0.5 μL of a mixture of dNTP (10 mM of dATP, dCTP, dGTP and dTTP), 1 μL of the primers (20 μM), 1 μL of polymerase enzyme GoTaq® (5U.μL-1, Promega®, Madison, WI, USA) and 16.5 μL DEPC-treated water. The PCR reaction was performed following González Salgado et al. (2011). PCR products were revealed by agarose gel electrophoresis, visualized in UV transilluminator, with previous staining in GelRedTM Biotium solution (2.5 ng. μL-1) and confirmed by the expected band size of 500 pb. Molecular size of the DNA fragments was estimated using the “qLadder 100 pb precision” marker (PB-L®, Bs As, Argentina).
Sclerotia production
Each isolate was cultured in 6-cm Petri dishes containing Czapeck dox (Cz) medium. They were inoculated in the center of the dish with 10 μL of a spore suspension and incubated for 14 days at 30°C (64). Sclerotia were recovered, their diameters measured under a microscope (Nikon eclipse Cs1 spectral) and characterized following Cotty (1989) into L or S morphotypes or isolates not producing (NP) sclerotia (60). Isolates not producing sclerotia after an incubation period were cultured in 5/2 medium for 5 to 7 days at 31°C to induce their production (38).
Toxigenic capability
Considering Aflatoxin B1 (AFB1) was one of the greatest frequency and toxicity of the four most important aflatoxins identified, its production was quantified (23,82). A. flavus isolates were inoculated in duplicate on MEA slants for 7 days at 28°C. These cultures were used to prepare spore suspensions, following the method described by Alaniz Zanon et al. (2013). Spore concentration was measured in a Neubauer chamber and adjusted to 105 spores. mL-1. Four-milliliter vials containing 1 mL broth from a medium containing 150 g sucrose, 20 g yeast extract, 10 g soytone, and 1 L distilled water, were inoculated with 100 μL of each spore suspension. Medium pH was adjusted to 5.9 with HCl. Cultures were incubated for 7 days at 30°C. Vial cultures were extracted according to Horn et al. (1996), by adding 1 mL of chloroform to each vial and vortexing for 30 s.
The first group of dried extracts was evaluated in their capacity to produce aflatoxins by HPLC according to Horn et al. (1996). Aflatoxins were analyzed by injecting 50 μL of extract from each vial into an HPLC system consisting of a Hewlett Packard model 1100 pump (Palo Alto, CA) connected to a Hewlett Packard model 1046A programmable fluorescence detector and a data module Hewlett Packard Kayak XA (HP ChemStation Rev.A.06.01). Chromatographic separations were performed on a stainless steel, C18 reversed-phase column (150 mm × 4.6 mm i.d., 5 μm particle size; Luna Phenomenex, Torrance, CA, USA) connected to a precolumn Security Guard (20 mm × 4.6 mm i.d., 5 μm particle sizes, Phenomenex). The mobile phase was water:methanol:acetonitrile (4:1:1, v/v/v) at a flow rate of 1.5 mL. min-1. Pure aflatoxin solutions were used as external standards (Sigma-Aldrich, St. Louis, MO, USA).
The second group of dried extracts was resuspended in 500 μL of mobile phase of acetonitrile: ZnSO4 4 mM buffer solution (65:35, V/V) and CPA was determined in the HPLC system. Chromatographic separations were performed in Agilent ZORBAX RX-SIL column (250 mm x 4.6 mm i.d., 5 μm particle size) connected to a pre-column Security Guard (20 mm x 4.6 mm i.d., 5 μm particle sizes, Phenomenex) and 50 μl of each simple were analyzed at a flow velocity of 0.8 mL. min-1.
The quantitative analysis was performed by normalization of peak areas. A calibration curve was elaborated from the areas obtained for the different concentrations of standards of aflatoxins and CPA, as applicable (Sigma Aldrich, St. Louis, MO, USA). The AFB1 and CPA detection limit was 1 ng. g-1 (24,32).
Sequencing and Phylogenetic relationships
Molecular variability of the non-aflatoxigenic isolates was explored using a 688-pb fragment of the CaM gene. We studied only this type of isolate given their potential for biocontrol strategies. Spores of the isolates cultivated in MEA medium were suspended for 7 days at 25°C. Conidia (1x106 conidia. mL-1) were inoculated in 100 mL of lixiviated medium of potato glucose agar (10% of potato infusion from 200 g potato, 2% glucose, 4.5 pH). They were incubated in orbital shaker at 150 rpm for 48 hours at 25°C. Mycelium was collected by filtration and powdered with liquid N2 (28). This culture medium allows one to obtain enough amount of mycelium. DNA was extracted using the cetyl-trimethylammonium bromide (CTAB) method (54), quantified by spectrophotometry and a segment of the CaM was amplified with the primers CL1, 5’- GA(GA)T(AT)CAAGGAGGCCTTCTC -3’; CL2A, 5’- TTTTTGCATCATGAGTTGGAC -3’ (61). The obtained fragments of the expected size (688 pb) were purified using DNA columns of Wizard® SV Gel columns and PCR clean - Up system (Promega, Madison, WI, USA), following manufacturer instructions. Then, they were quantified by spectrophotometry. Both DNA chains were sent for sequencing by Sanger method (Macrogen, Seoul, South Korea). The obtained sequences were aligned using the software Clustal X2 (54). The software MEGA version 7 (53) was used to select the best nucleotide substitution model and to obtain the phylogenetic trees using Neighbor-Joining (NJ) (74) and Maximum-Likelihood (ML) methods (39,76), both with 10,000 replicates bootstrap. The sequences were compared with the reference NW_002477238.1 and A. niger MH645004.1 was used as outgroup, both from the GenBank.
Statistical analysis
Data on sclerotia morphotypes, toxigenic capacity and variability of the sequences of the CaM genes were analyzed using InfoStat statistical software as well as correlation analyses (29). Mean values were obtained using analysis of variance (ANOVA) and differences between means were compared using Fisher’s LSD test (P< 0.05).
Results and discussion
Morphological and molecular identification
A total of 58 isolates were obtained in the sampling localities from the eight agro-climatic zones (table 1, page 63-64) with A. flavus morphological characteristics, according to Klich (2002) (figure 2, page 64).
Table 1 Isolates of Aspergillus flavus from maize ears collected from representative localities of eight agro-climatic zones of the Chaco Semi-arid region of Argentina. Production of aflatoxin B1, cyclopiazonic acid and sclerotia morphotype during the 2015/16 and 2016/17 growing seasons. Tabla 1. Aislados de Aspergillus flavus de espigas de maíz colectadas en localidades representativas de ocho zonas agroclimáticas del Chaco semiárido argentino. Producción de aflatoxina B1, ácido ciclopiazónico y morfotipo de esclerocios durante las campañas agrícolas 2015/16 y 2016/17.
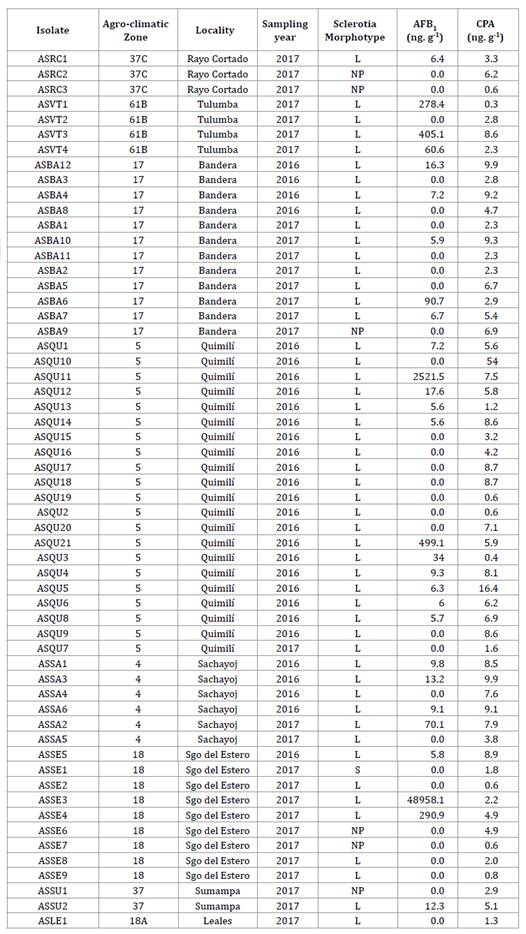
Agro-climatic zones: 4: Río Muerto, 5: Hickmann, 17: Bandera, 18 and 18A: Monteagudo, 37: Soto, 37C: Soto - North of Córdoba, 61B: Dean Funes. L: sclerotia of > 400 μm; S: sclerotia of < 400 μm; NP: non-producer of sclerotia. AFB1: Aflatoxin B; CPA: cyclopiazonic acid.
Zonas agroclimáticas: 4: Río Muerto, 5: Hickmann, 17: Bandera, 18 y 18A: Monteagudo, 37: Soto, 37C: Soto - Norte de Córdoba, 61B: Deán Funes. L: esclerocios de tamaño > 400 μm; S: esclerocios de tamaño < 400 μm; NP: no productor de esclerocios. AFB1: Aflatoxina B; CPA: ácido ciclopiazónico.

Figure 2 Aspergillus flavus Link colony on malt extract agar (MEA) culture medium. Figura 2. Colonia de Aspergillus flavus Link en medio de cultivo agar extracto de malta (MEA).
A. flavus identity was confirmed using the specific primers FLA1 and FLA2 through amplification of a 500-pb band (41) (figure 3, page 65).
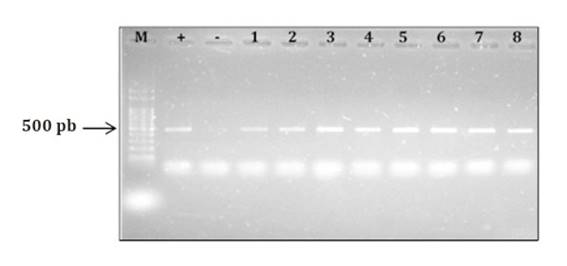
M: molecular marker (100-pb DNA Ladder); +: Aspergillus flavus NRRL 3251 (positive control); -: A. parasiticus NRRL2999 (negative control); 1: ASSA2; 2: ASQU7; 3: ASQU21; 4: ASQU10; 5: ASQU15; 6: ASRC2; 7: ASQU12; 8: ASQU6; 9: ASBA3; 10: ASVT1.
M: marcador molecular (100-pb DNA Ladder); +: Aspergillus flavus NRRL 3251 (control positivo); -: A. parasiticus NRRL2999 (control negativo); 1: ASSA2; 2: ASQU7; 3: ASQU21; 4: ASQU10; 5: ASQU15; 6: ASRC2; 7: ASQU12; 8: ASQU6; 9: ASBA3; 10: ASVT1.
Figure 3 Electrophoresis gel showing PRC products obtained using the specific primers FLA1 and FLA2 (González Salgado et al., 2011). Figura 3. Gel de electroforesis mostrando productos de PCR obtenidos utilizando los iniciadores específicos FLA1 y FLA2 (González Salgado et al., 2011).
The presence of isolates in the eight zones, each one being considered a different agroecological environment given different meteorological scenarios at each growing season shows the broad distribution of the fungus in the study region, as reported for other regions of the country (8,17) and the world (11,30,32).
Sclerotia production
Of the 58 A. flavus isolates identified, 6 (10%) produced no sclerotia, whereas 52 (90%) produced sclerotia. Of the latter, 51 (98%) were identified as L morphotype (figure 4), with ˃ 400 μm in diameter, and only isolate, ASSE1, (2%) corresponded to S morphotype, with ˂ 400 μm in diameter (table 1, page 63-64).
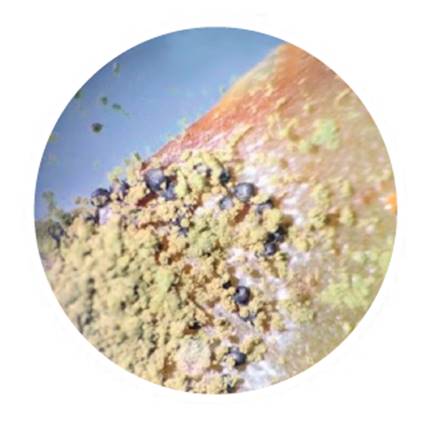
Figure 4 Sclerotia L morphotype in aflatoxigenic isolate of Aspergillus flavus. Observation under stereo microscope Nikon SMZ-10 (15X). Figura 4. Esclerocios morfotipo L en aislado aflatoxigénico de Aspergillus flavus. Observación bajo lupa estereoscópica Nikon SMZ-10 (15X).
Similar proportions between L, S and NP morphotypes in maize kernels were reported by Moreno (2004) in Mexico and by Pildain et al. (2005) in Argentina in peanut isolates. The low or null presence of S morphotype in maize was also reported by Giorni et al. (2007) in Italy. These authors identified one S isolate out of 70 isolates, with the remaining ones being NP. Similarly, Alaniz Zanon et al. (2018) reported a high proportion of L morphotype in Argentina with respect to NP, with absent S morphotype.
Sclerotia production was reported in isolates from all the agro-climatic zones and in both evaluated growing seasons. Dominance of L morphotype among isolates obtained in the Chaco Semi-arid region agrees with previous records in Argentina (7,18) and in other countries, like Brazil, Portugal, Nigeria and Sub-Saharan Africa regions (5,20,33,68,72).
The high incidence of S morphotype is associated with regions of low precipitation and high temperatures, where high number of small sclerotia can be a survival trait facing rapid temperature and humidity fluctuations (19). However, and even though the study region is dry and hot, this was not observed.
Toxigenic capability
Based on the production of AFB1 and CPA of the 58 isolates obtained, we identified 30 (52%) non-aflatoxigenic and 28 (48%) aflatoxigenic. This similarity of proportions was indicated by Martins et al. (2017) in peanut crop in Brazil and Camiletti et al. (2018) in maize ears in Argentina.
Aflatoxin-producer and non-producer isolates were obtained in all the agro-climatic zones and growing seasons, except Santa Rosa de Leales, where the only isolate found was non-producer.
Of the 28 isolates producing AFB1, 26 (93%) had concentrations below 500 ng. g-1, whereas ASQU11 and ASSE3 were higher aflatoxin producers of 2,521.5 ng. g-1 and 48,958.1 ng. g-1 respectively (table 1, page 63-64). Although the capacity to produce mycotoxins may depend on geographical origin and environmental conditions (62), our results do not show any pattern associated with zones or growing seasons. This agrees with Bayman and Cotty (1993), who did not detect differential patterns between nearby zones.
All isolates were CPA producers, indicating toxicity importance (78) and plausible use as aflatoxin biocontrol strategies. This is the case of the AF36 agent used in competitive exclusion strategies and found responsible for CPA increase in maize kernels inoculated in the field, and peanuts (2,31). By contrast, Camiletti et al. (2018) detected 19% of CPA non-producer isolates in maize kernels in Argentina. On the other hand, Vaamonde et al. (2003) studied wheat, soybean and peanut in Argentina and found between 6% and 27% of isolates not producing this mycotoxin. In Italy, Giorni et al. (2007) reported 39% CPA non-producer isolates in maize kernels, whereas, in organic nut plantations in Brazil, Reis et al. (2014) found that 34% of the A. flavus isolates were non-producers of that mycotoxin. In addition, Jamali et al. (2012) identified 19 % of the isolates as CPA non-producers in soil samples of pistachio plantations in Czech Republic.
The presence of isolates producing both CPA and aflatoxins was reported in Argentina and other parts of the world (8,18,37,71). The isolates with the highest AFB1 production obtained in the Chaco Semi-arid region do not correspond to those with the highest CPA production.
Correlations between AFB1 production and the number (r= 062) and size of sclerotia (r= -0.38) indicate that the produced amount of AFB1 increases as sclerotia size decreases. These results are similar to those reported by Arrúa Alvarenga et al. (2012) and Pildain et al. (2005), but differ from Bouti et al. (2020), who did not find any correlation between sclerotia and aflatoxin production. No correlation was observed between CPA production and sclerotia morphotype.
The only S morphotype identified in this study (ASSE1), collected in 2016/17 growing season, did not produce AFB1. While the presence of non-aflatoxin-producing isolates was also reported for USA, Ghana and Brazil (3,22,40,44), in general, S morphotype isolates produce higher aflatoxin concentrations than L or NP morphotypes and are identified as major causal agents of severe contamination in maize and of most human deaths due to aflatoxicosis (13,23). The S isolates are worldwide distributed, associated with aflatoxins B production in the USA and Africa (68), and of both B and G aflatoxins (3,38).
Sequencing and phylogenetic analysis
All the CaM gene sequences studied in this work, using both NJ and ML, belong to the A. flavus clade (figure 5, page 67 and figure 6, page 68).

Node number corresponds to bootstrap support values (10000 replicates). A. flavus NW 002477238.1 was selected as reference sequence and A. niger MH645004.1 as outgroup sequence.
El número de nodos corresponde a un valor de bootstrap de 10000 réplicas. A. flavus NW 002477238.1 se utilizó como secuencia de referencia y A. niger MH645004.1 como secuencia externa al grupo.
Figure 5 Phylogenetic tree built using the Neighbor-Joining (NJ) statistic method based on the relationship among segments of the sequence of the CaM gene from A. flavus isolates from agro-climatic zones of the Chaco Semi-arid region in Argentina. Figura 5. Árbol filogenético realizado con el método estadístico Neighbor-Joining (NJ) basado en las relaciones entre las secuencias de segmentos del gen CaM de aislados de A. flavus provenientes de zonas agroclimáticas del Chaco semiárido argentino.

Node number corresponds to bootstrap support values (10000 replicates). A. flavus NW 002477238.1 was selected as reference sequence and A. niger MH645004.1 as outgroup sequence.
El número de nodos corresponde a un valor de bootstrap de 10000 réplicas. A. flavus NW 002477238.1 se utilizó como secuencia de referencia y A. niger MH645004.1 como secuencia externa al grupo.
Figure 6 Phylogenetic tree built using the Maximum-Likelihood (ML) statistical method based on the relationship among segments of sequences of the CaM gene from A. flavus isolates from agro-climatic zones in the Chaco Semi-arid region in Argentina. Figura 6. Árbol filogenético realizado con el método estadístico Maximum-Likelihood (ML) basado en las relaciones entre las secuencias de segmentos del gen CaM de aislados de A. flavus provenientes de zonas agroclimáticas del Chaco semiárido argentino.
The high similarity among isolates for that character showed limited genetic diversity in this geographic region. In addition, the only S morphotype is located together with L morphotype isolates, all of which are aflatoxin non-producers. This finding may be attributed to the fact that these are young populations and, therefore, may have not undergone sufficient mutations or recombination events leading to variability among isolates from the region (42,62). However, only the northern zones of the region (4 and 5) correspond to land recently converted to agriculture, whereas in the remaining districts, both commercial hybrids and maize for self-consumption have been cultivated for several decades.
Conclusions
A. flavus is present in all localities of the sampled agro-climatic zones and both growing seasons, evidencing the wide distribution of the pathogen in the Chaco Semi-arid region. The population of A. flavus exhibits diversity in terms of sclerotia morphotypes and production of AFB1 and CPA.
Low genetic variability among the isolates was observed, all belonging to the A. flavus clade.
In the future, genes from more variable regions will be used to perform phylogenetic analyses among A. flavus isolates.